Copyright
©The Author(s) 2018.
World J Gastroenterol. Dec 14, 2018; 24(46): 5223-5233
Published online Dec 14, 2018. doi: 10.3748/wjg.v24.i46.5223
Published online Dec 14, 2018. doi: 10.3748/wjg.v24.i46.5223
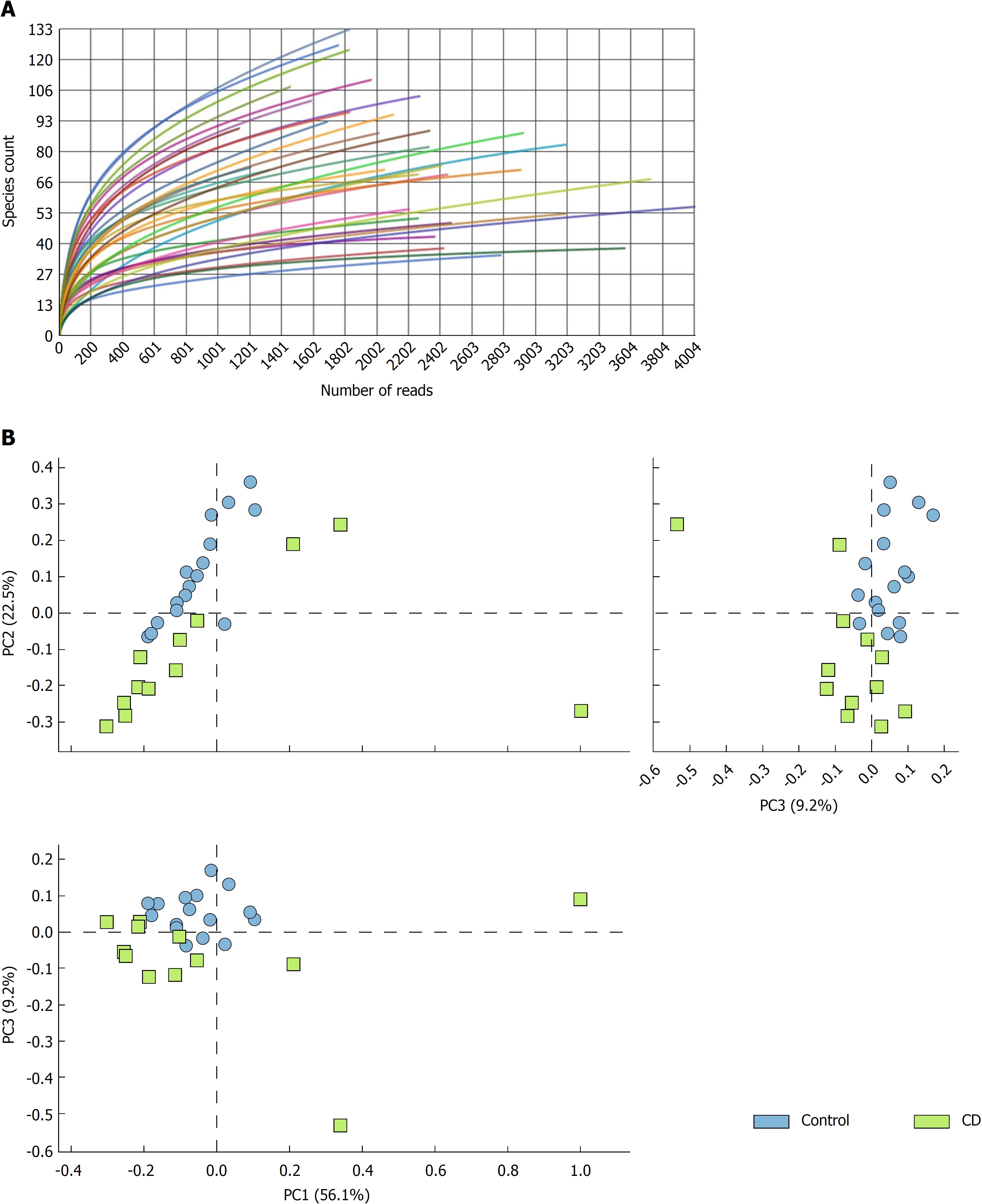
Figure 1 Ecological and metagenomic analysis.
A: Number of readings for each sample using the 16S massive sequencing technique in GS Junior. See methods section for details. B: Principal components analysis. The control samples and Crohn’s disease are observed in well-defined groups. The data was selected with the Ribosomal Project database using a maximum e-value of 10-5, a minimum identity of 75%, and a minimum length alignment of 15 bp. PC: Principal components; CD: Crohn’s disease sample.
- Citation: Rojas-Feria M, Romero-García T, Fernández Caballero-Rico JÁ, Pastor Ramírez H, Avilés-Recio M, Castro-Fernandez M, Chueca Porcuna N, Romero-Gόmez M, García F, Grande L, Del Campo JA. Modulation of faecal metagenome in Crohn’s disease: Role of microRNAs as biomarkers. World J Gastroenterol 2018; 24(46): 5223-5233
- URL: https://www.wjgnet.com/1007-9327/full/v24/i46/5223.htm
- DOI: https://dx.doi.org/10.3748/wjg.v24.i46.5223