Published online Aug 14, 2018. doi: 10.3748/wjg.v24.i30.3347
Peer-review started: April 10, 2018
First decision: May 16, 2018
Revised: May 29, 2018
Accepted: June 25, 2018
Article in press: June 25, 2018
Published online: August 14, 2018
Processing time: 125 Days and 9.8 Hours
The clinical outcome of Hepatitis B Virus (HBV) infection depends on the success or failure of the immune responses to HBV, and varies widely among individuals, ranging from asymptomatic self-limited infection, inactive carrier state, chronic hepatitis, cirrhosis, hepatocellular carcinoma, to liver failure. Genome-wide association studies (GWAS) identified key genetic factors influencing the pathogenesis of HBV-related traits. In this review, we discuss GWAS for persistence of HBV infection, antibody response to hepatitis B vaccine, and HBV-related advanced liver diseases. HBV persistence is associated with multiple genes with diverse roles in immune mechanisms. The strongest associations are found within the classical human leukocyte antigen (HLA) genes, highlighting the central role of antigen presentation in the immune response to HBV. Associated variants affect both epitope binding specificities and expression levels of HLA molecules. Several other susceptibility genes regulate the magnitude of adaptive immune responses, determining immunity vs tolerance. HBV persistence and nonresponse to vaccine share the same risk variants, implying overlapping genetic bases. On the other hand, the risk variants for HBV-related advanced liver diseases are largely different, suggesting different host-virus dynamics in acute vs chronic HBV infections. The findings of these GWAS are likely to pave the way for developing more effective preventive and therapeutic interventions by personalizing the management of HBV infection.
Core tip: Genome-wide association studies (GWAS) have proven to be very useful in uncovering the host genetic factors that influence the clinical outcomes of hepatitis B virus (HBV) infection. Both class I and class II human leukocyte antigen (HLA) genes were implicated in persistence of HBV infection; associated variants affected antigen-binding specificities and expression levels of HLA molecules. HBV persistence and vaccine nonresponse were associated with the same HLA-DP allotypes, suggesting a critical role for the surface antigen in HBV pathogenesis. These findings might be exploited for development of potent vaccines based on alternative epitopes. GWAS for HBV-related pathologies identified many other immune-related genes, and provided genetic markers to detect the individuals at high risk for HBV-related diseases.
- Citation: Akcay IM, Katrinli S, Ozdil K, Doganay GD, Doganay L. Host genetic factors affecting hepatitis B infection outcomes: Insights from genome-wide association studies. World J Gastroenterol 2018; 24(30): 3347-3360
- URL: https://www.wjgnet.com/1007-9327/full/v24/i30/3347.htm
- DOI: https://dx.doi.org/10.3748/wjg.v24.i30.3347
Hepatitis B virus (HBV) is the most common viral pathogen of the human liver, and is a prominent cause of acute and chronic hepatitis, liver failure, cirrhosis and hepatocellular carcinoma (HCC). Around 257 million people, or 3.5% of the global population, are estimated to have chronic HBV infection[1], and more than 800 thousands people lose their life annually due to HBV-related complications[2]. Perinatal and childhood infections are very common in regions with high endemicity, and mostly result in life-long persistence, whereas infections at adulthood are mostly self-limited[3]. Chronic HBV carriers are at a high risk of developing end-stage liver diseases, such as cirrhosis and HCC[4]. Indeed, HBV infection is responsible for 27% of cirrhosis and 53% of HCC cases worldwide[5]. Hepatitis B vaccine effectively prevents new infections, while antiviral medicines suppress progression of HBV-related liver damage. However, vaccination, safe healthcare practices, and access to treatment do not have full population coverage, and HBV infection still remains a major public health problem[6].
HBV is generally considered to be non-cytopathic per se; the liver injury associated with HBV infection is immune-driven. The clinical outcome of HBV infection varies greatly among individuals, ranging from asymptomatic self-limited infection, inactive carrier state, chronic hepatitis to end-stage liver diseases with life-threatening complications[3,7-9]. Viral, host and environmental factors affect these outcomes. Viral core/pre-core mutations, certain viral genotypes (e.g., genotype C in comparison to genotype B), high viral load, early-life (perinatal and childhood) infections, male sex, host genetic factors (e.g., HLA class II homozygosity), suppressed immune status, co-existing metabolic diseases, and exposure to hepatotoxic substances (e.g., aflatoxin, alcohol) are associated with worse prognosis[7,10]. A complete understanding of these factors is crucial for developing more tailored and effective preventive and therapeutic interventions to reduce the burden of HBV-related complications.
The contribution of host genetic factors to the variation in HBV infection outcomes was most notably evidenced by twin studies[11], which reported higher concordance rates in monozygotic twins than in same-sex dizygotic twins for HBV carrier status[12] and for antibody titers in response to hepatitis B vaccine[13,14]. The host genetic factors were investigated in association studies whereby the frequencies of genetic variants were compared between case and control groups, using candidate gene and whole genome approaches[15-18]. Candidate gene studies focused on polymorphisms in immunologically relevant genes, especially the classical human leukocyte antigen (HLA) genes[19-21]. HLA genes encode the molecules that present antigens to T lymphocytes, and polymorphisms in these genes may alter the specificity and strength of antigen binding, affecting the T cell-mediated immune responses. In accordance with this paradigm, HLA typing studies found an ample amount of HLA allelic variations associated with the clinical outcomes of HBV infection[15]. However, the associations reported in these studies were largely inconsistent, even within the same ethnicity, with few exceptions. The validity of these studies were undermined by inappropriately small sample sizes, lack of replication in an independent cohort, ambiguous allele assignments, genotyping confined to only one or two exons that show the highest genetic variability, low population coverage, and weak statistical evidences[11,15].
The development of high-throughput genotyping technologies (e.g., microarrays) and the construction of a detailed map of common genetic polymorphisms in humans enabled genome-wide investigation of genetic variants for association to complex traits and diseases. In contrast to candidate gene-based studies, genome-wide association studies (GWAS) test hundreds of thousands to millions of common SNPs across the genome, providing an unbiased method to investigate genetic risk loci, and allowing the discovery of novel disease-relevant genes. Several GWAS were conducted to identify the risk loci that predisposes to persistence of HBV infection, non-response to hepatitis B vaccine, and progression of liver disease in chronic HBV infections. In this review, we discuss the findings of these GWAS, and we emphasize how GWAS has driven the research on the genetic basis of variability in HBV-related pathologies.
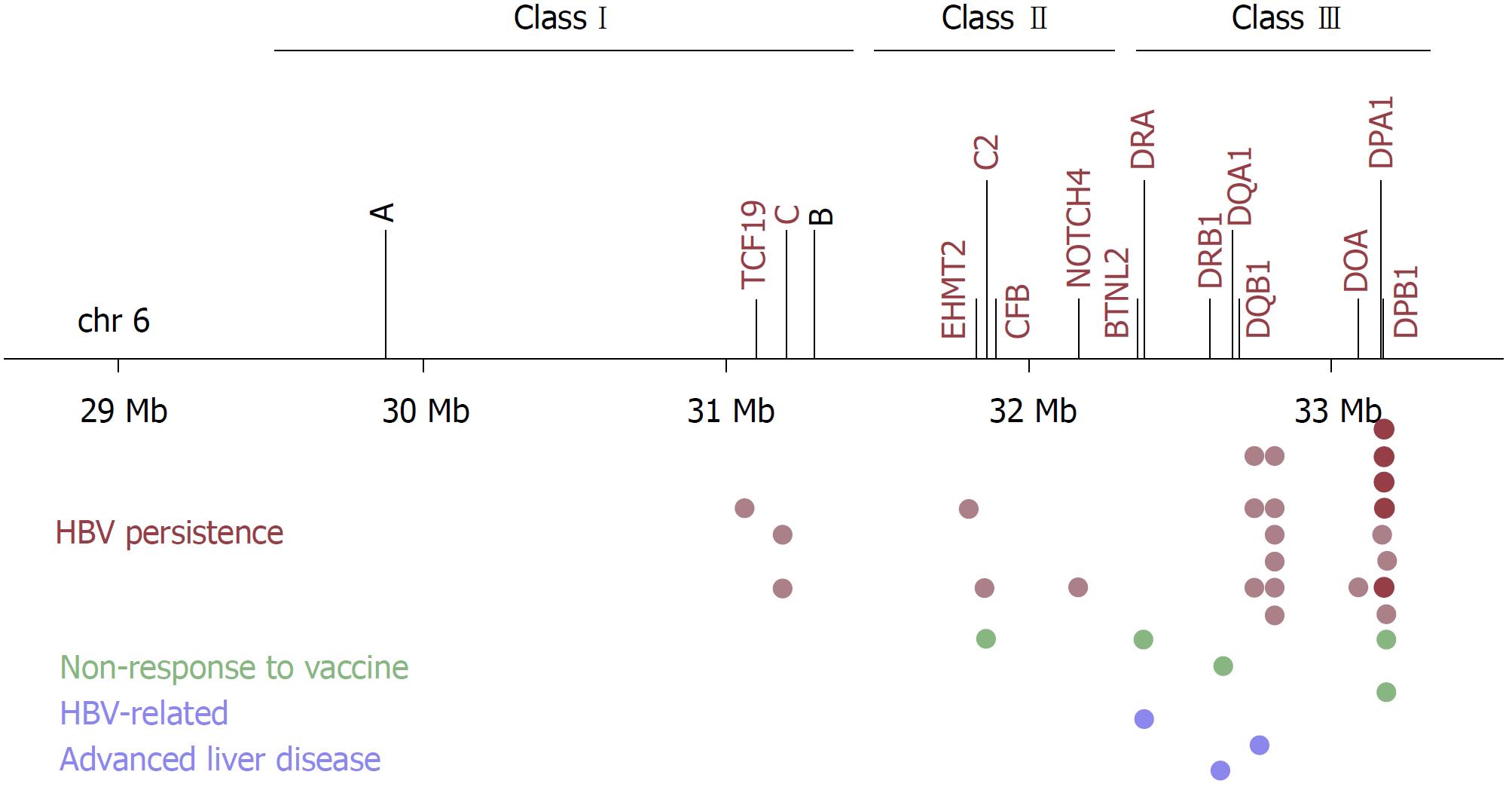
The first GWAS to identify the genetic risk factors for susceptibility to HBV infection persistence was performed in a Japanese population, and published in 2009[22]. Seven hundred and eighty-six chronic hepatitis B (CHB) cases and 2201 HBsAg seronegative controls were used in the discovery phase. This GWAS detected significantly associated SNPs within the HLA-DP locus. rs3077 in HLA-DPA1 3’UTR and rs9277535 in HLA-DPB1 3’UTR were selected, and further replicated in independent Japanese and Thai samples[22]. The same group employed a second GWAS using additional Japanese case-control samples where they confirmed the associations of HLA-DP variants, and, additionally, detected significant associations around the HLA-DQ locus[23]. The HLA-DQ SNPs, rs2856718 and rs7453920, were not in LD with the HLA-DP SNPs, and their associations remained significant after adjusting for the effect of rs9277535. These findings revealed that HLA-DP and HLA-DQ variants were independently associated with chronic HBV infection. rs2856718 tagged an LD block comprising HLA-DQA1 and HLA-DQB1, which produce the functional HLA-DQ molecules. rs7453920 was located in HLA-DQB2, which, along with HLA-DQA2, is thought to be nonfunctional. The association of rs7453920 with functional HLA-DQ alleles was shown later[24].
Additional GWAS for HBV persistence were performed in Japanese[25], Korean[26], Han Taiwanese[27], and Han Chinese populations[28-30] (Table 1, Figure 1). These GWAS repeatedly mapped the strongest signals within the HLA-DP and/or HLA-DQ loci, replicating the associations of rs3077, rs9277535, rs7453920 and rs2856718. Furthermore, independent studies genotyping the same SNPs validated these associations with high consistency in diverse Asian populations, including Chinese[31-39], Japanese[40], Thai[41], Indonesians[42], Tibetans and Uygurs (Asian-Caucasian mix)[43]. A meta-analysis of 62050 subjects from 29 case-control studies, mostly from Asian populations, further validated the associations of rs3077 and rs9277535[44]. Replication studies were also conducted in non-Asian populations, including Caucasians in Germany[45], European Americans, African Americans[46], Saudi Arabians population[47], and multiethnic Argentine populations (Native American-European Caucasian mix)[48]. With few exceptions and inconsistencies, probably due to different allele frequencies and LD structures in these populations, the associations of HLA-DP (rs3077 and rs9277535) and HLA-DQ (rs7453920 and rs2856718) SNPs were validated in these populations, too.
| Ref. | Studypopulation | SNPID | Minor/major alleles | Riskallele | P | OR | Location | Nearestgene | Functional class |
| Kamatani et al[22] | Japanese, Thai (n = 6387)1 | rs3077 | A/G | G | 2.31 × 10-38 | 0.56 | 6p21.32 | HLA-DPA1 | 3'UTR |
| rs9277535 | A/G | G | 6.34 × 10-39 | 0.57 | 6p21.32 | HLA-DPB1 | 3'UTR | ||
| Mbarek et al[23] | Japanese (n = 9163)1 | rs3077 | A/G | G | 1.57 × 10-61 | 1.87 | 6p21.32 | HLA-DPA1 | 3'UTR |
| rs9277535 | A/G | G | 2.55 × 10-54 | 1.77 | 6p21.32 | HLA-DPB1 | 3'UTR | ||
| rs2856718 | A/G | A | 3.99 × 10-37 | 1.56 | 6p21.32 | HLA-DQB1, HLA-DQA2 | Intergenic | ||
| rs7453920 | A/G | G | 5.98 × 10-28 | 1.81 | 6p21.32 | HLA-DQB2 | Intron | ||
| Nishida et al[25] | Japanese (n = 1793)2 | rs3077 | A/G | G | 4.40 × 10-19 | 0.46 | 6p21.32 | HLA-DPA1 | 3'UTR |
| rs9277542 | A/G | G | 1.28 × 10-15 | 0.5 | 6p21.32 | HLA-DPB1 | Exon | ||
| Kim et al[26] | Korean (n = 4309)2 | rs3077 | A/G | G | 3.74 × 10-40 | 0.53 | 6p21.32 | HLA-DPA1 | 3'UTR |
| rs9277535 | A/G | G | 5.25 × 10-39 | 0.53 | 6p21.32 | HLA-DPB1 | 3'UTR | ||
| rs2856718 | A/G | A | 1.78 × 10-24 | 1.6 | 6p21.32 | HLA-DQB1, HLA-DQA2 | Intergenic | ||
| rs7453920 | A/G | G | 6.71 × 10-26 | 0.5 | 6p21.32 | HLA-DQB2 | Intron | ||
| rs652888 | G/A | G | 7.07 × 10-13 | 1.38 | 6p21.33 | EHMT2 | Intron | ||
| rs1419881 | G/A | A | 1.26 × 10-18 | 0.73 | 6p21.33 | TCF19 | 3'UTR | ||
| Hu et al[28] | Chinese (n = 11791)3 | rs7453920 | A/G | G | 4.93 × 10-37 | 0.53 | 6p21.32 | HLA-DQB2 | Intron |
| rs3130542 | A/G | A | 9.49 × 10-14 | 1.33 | 6p21.33 | HLA-C | Intergenic | ||
| rs4821116 | A/G | G | 1.71 × 10-12 | 0.82 | 22q11.21 | UBE2L3 | Intron | ||
| rs3077 | A/G | G | 6.50 × 10-14 | 0.75 | 6p21.32 | HLA-DPA1 | 3'UTR | ||
| Chang et al[27] | Han Taiwanese (n = 2688)2 | rs9277535 | A/G | G | 4.87 × 10-14 | 1.59 | 6p21.32 | HLA-DPB1 | 3'UTR |
| rs7453920 | A/G | G | 6.66 × 10-15 | 2.31 | 6p21.32 | HLA-DQB2 | Intron | ||
| Jiang et al[29] | Chinese (n = 18371)1 | rs12614 | T/C | C | 1.28 × 10-34 | 1.89 | 6p21.33 | CFB | Exon |
| rs422951 | G/A | A | 5.33 × 10-16 | 1.27 | 6p21.32 | NOTCH4 | Exon | ||
| rs378352 | T/C | T | 1.04 × 10-23 | 1.26 | 6p21.32 | HLA-DOA | Exon | ||
| rs2853953 | A/G | G | 5.06 × 10-20 | 1.47 | 6p21.33 | HLA-C | Intergenic | ||
| rs1883832 | T/C | T | 2.95 × 10-15 | 1.19 | 20q13.12 | CD40 | 5'UTR | ||
| rs2856718 | G/A | A | 7.35 × 10-28 | 1.28 | 6p21.32 | HLA-DQB1, HLA-DQA2 | Intergenic | ||
| rs7453920 | A/G | G | 1.28 × 10-60 | 2 | 6p21.32 | HLA-DQB2 | Intron | ||
| rs9277535 | A/G | G | 9.84 × 10-71 | 1.52 | 6p21.32 | HLA-DPB1 | 3'UTR | ||
| rs3077 | A/G | G | 1.15 × 10-53 | 1.45 | 6p21.32 | HLA-DPA1 | 3'UTR | ||
| Li et al[30] | Chinese (n = 9569)3 | rs7000921 | C/T | T | 3.20 × 10-12 | 0.78 | 8p21.3 | INTS10 | Intergenic |
| rs7453920 | A/G | G | 6p21.32 | HLA-DQB2 | Intron | ||||
| rs9277535 | A/G | G | 6p21.32 | HLA-DPB1 | 3'UTR |
Most of the subsequent GWAS also revealed additional susceptibility loci for HBV persistence. To begin with, a GWAS in Korean chronic HBV carriers and non-infected healthy individuals identified two novel loci within the HLA region[26]. These two loci, marked by rs652888 in EHMT2 and rs1419881 in TCF19, had independent effects on HBV persistence. EHMT2 is a histone lysine methyltransferase involved in gene expression regulation, and plays roles in immune cell development and differentiation[49]. TCF19 is a transcription factor necessary for cell survival and proliferation[50]. A previous GWAS associated a non-synonymous SNP in TCF19 with blood cell counts, including lymphocyte and monocyte (macrophage precursor) cell counts[51], suggesting a potential mechanism by which TCF19 variants affected immune mechanisms. However, the associations of rs652888 (EHMT2) and rs1419881 (TCF19) were not replicated in Chinese and Thai populations, probably due to different genetic structures[24,30,41]. Additional replication studies are, therefore, necessary for both loci.
Another GWAS in Han Chinese used chronic HBV carriers as cases and individuals who naturally cleared HBV infection as controls, and revealed two additional loci[28]. The first locus was marked by the lead SNP rs3130542, located near HLA-C within the HLA region. Conditional analysis showed that the effect of rs3130542 was independent of HLA-DP and HLA-DQ variants. Therefore, both class I and class II variants were detected by GWAS for HBV persistence. The second locus was marked by the lead SNP rs4821116, located in UBE2L3 on chromosome 22. UBE2L3 encodes an ubiquitin-conjugating enzyme, which enhances NFκB activation upon CD40 stimulation in B cells and TNF stimulation in monocytes[52]. The protective variant of rs4821116 was associated with higher UBE2L3 mRNA levels in peripheral blood monocytes[53]. Moreover, UBE2L3 was implicated in multiple autoimmune diseases; the risk variants correlated with higher UBE2L3 expression in B cells and monocytes, enhanced NFκB activation, enhanced B cell proliferation and activation[52]. Thus, hyperactivity of UBE2L3-related immune mechanisms conferred protection from infections on the one hand, but predisposed to autoimmunity on the other hand.
The GWAS with the largest sample size (2514 chronic HBV carriers and 1130 healthy controls of Chinese ethnicity in the discovery phase) was published in 2015, revealing five new SNPs independently associated with HBV persistence[29]. Four of these SNPs, rs12614 in CFB, rs422951 in NOTCH4, rs2853953 near HLA-C, and rs378352 in HLA-DO were located within the HLA region, whereas the other SNP, rs1883832, was located in CD40 on chromosome 20. rs12614_T/C in CFB represents a non-synonymous amino acid polymorphism (R32W) in complement factor B. The complement system is part of the innate immune response against viral infections, and is also involved in enhancing adaptive immune responses[54]. Chronic HBV carriers and individuals with the risk genotype CC for rs12614 had significantly lower plasma CFB protein levels[29]. rs422951 in NOTCH4 also causes a non-synonymous amino acid change (T320A). Notch signaling regulates immune cell development and T-cell mediated immune responses[55]. rs2853953 tagged HLA-C gene; however it was not in LD with the previously reported HLA-C tagging SNP rs3130542. Indeed, these two SNPs marked different HLA-C alleles; rs2853953 marked HLA-C*06:02, whereas rs3130542 marked HLA-C*07:02. rs378352 is located in exon 4 of HLA-DOA, causing a synonymous codon change. HLA-DOA encodes the α chain of the HLA-DO molecule, which, along with HLA-DM, regulates peptide loading to class II molecules. Whether or how this polymorphism affects HLA-DO function is currently unknown. Lastly, rs1883832_T/C is located in the Kozak sequence of CD40, and was previously shown to affect its translational efficiency[56]. The risk genotype TT for rs1883832 correlated with lower CD40 levels in the serum in both cases and controls[29]. CD40 is a co-stimulatory receptor protein found on antigen-presenting cells. Its engagement with CD40 ligand (CD40L) on T cells initiates and promotes humoral and cellular adaptive immune responses[57]. Intriguingly, rs1883832_T was previously associated with protection against multiple autoimmune diseases[56]. Thus, CD40 plays an important role in regulating the magnitude of immune responses, and determining immunity vs tolerance. So far, the associations of rs12614 (CFB) and rs1883832 (CD40) were validated by independent studies in Han Chinese[24,30]. Additional validation and functional studies are required to establish the roles of other genes and genetic variants in HBV persistence.
The most recent GWAS identified another novel locus (tagged by rs7000921) on chromosome 8 using persistently infected cases and spontaneously recovered controls in a Chinese population[30]. rs7000921 was an eQTL for the nearby INTS10 gene; the protective allele was associated with elevated INTS10 expression in liver. INTS10 encodes a subunit of the Integrator complex, which has multiple roles in transcriptional regulation. Subsequent analysis showed that INTS10 was involved in suppressing HBV replication in hepatocyte cell lines, likely through an interferon-dependent mechanism[30].
To sum up, GWAS for HBV infection persistence revealed many candidate genes with diverse functions in immune responses, improving our understanding of the molecular mechanisms leading to the success or failure of HBV clearance.
GWAS association signals within the HLA-DP, HLA-DQ and HLA-C loci implicated variable efficiencies in presenting immunodominant HBV epitopes to T cells. Thus, in the next step after GWAS, the respective HLA genes were typed by direct sequencing or imputation (on the basis of typed SNPs from the study populations and haplotypes from the genetic reference populations), and classical HLA alleles with significant effects on HBV persistence were determined (Table 2). HLA-DP susceptible alleles (DPA1*02:02; DPB1*05:01, *09:01) and protective alleles (DPA1*01:03; DPB1*02:01, *04:01, *04:02) were identified with a high degree of consistency in multiple Japanese, Korean and Chinese populations[22,24,28,30,58,59]. Moreover, DPB1*01:01 and DPB1*04:01 were identified as the most susceptible and the most protective DPB1 allele, respectively, in European Americans and African Americans[46]. Alleles with consistent effects in more than one population were also found for HLA-DQ and HLA-C genes; DQB1*03:02 was revealed as a protective allele[24,28,30,59], whereas DQA1*06:01, DQB1*03:01, DQB1*06:01, and HLA-C*07:02 were revealed as susceptible alleles[23,24,28-30,59]. Systematic epitope discovery studies are warranted to identify the HBV-derived peptides efficiently presented by protective allotypes, but not by susceptible allotypes. The results of such studies might form the basis for epitope-specific therapeutic interventions.
| HLA gene | Associated alleles | Effect on HBV persistence | Study population | Ref. |
| HLA-C | *07:02 | Protective | Chinese | [24,28,29] |
| HLA-DPA1 | *01:03 | Protective | Japanese, Korean, Chinese, Thai | [22,24,58] |
| *02:02 | Susceptible | Japanese, Korean, Chinese | [22,24,58] | |
| HLA-DPB1 | *04:01 | Protective | Japanese, Chinese | [22,28,58] |
| *04:02 | Protective | Japanese, Korean | [22,58] | |
| *02:01 | Protective | Chinese, Japanese | [28,58] | |
| *05:01 | Susceptible | Japanese, Korean, Chinese | [22,24,58,59] | |
| *09:01 | Susceptible | Japanese, Chinese | [22,24,58,59] | |
| HLA-DQA1 | *06:01 | Susceptible | Chinese | [24,28,30] |
| HLA-DQB1 | *03:02 | Protective | Japanese, Chinese | [24,28,30,59] |
| *03:01 | Susceptible | African American, Chinese | [18,28,30] | |
| *06:01 | Susceptible | Japanese | [23,59] | |
| HLA-DRB1 | *13:02 | Protective | Japanese, Chinese, Gambian, Korean, Germany, European Americans | [24,59,68] |
A major distinction between DPB1 protective alleles (*02:01, *04:01, *04:02) and DPB1 susceptible alleles (*01:01, *05:01, *09:01) was in their amino acid residues at positions 84-87. Protective alleles encoded Gly/Gly/Pro/Met (GGPM), while susceptible alleles encoded Asn/Glu/Ala/Val (DEAV) at 84-87. These residues corresponded to the pocket-1 of the antigen-binding groove, an anchor region that determines the binding specificity of DP molecules[60,61]. Epitope binding studies revealed that DP molecules with 84GGPM87 strongly bound aromatic and hydrophobic residues in their hydrophobic pocket-1, whereas DP molecules with 84DEAV87 also preferred positively charged residues, owing to the acidic residues at 84 and 85[61,62]. These distinct binding motifs of DP allotypes likely form the basis of HBV chronicity as multiple risk-associated DP allotypes ineffectively present a common epitope motif and lead to the failure of viral elimination. The identities of HBV peptides with this motif and their immunogenic properties remain to be determined by further functional assays.
It should be noted that the contribution of HLA-DP allotypes to HBV chronicity might also involve previously unprecedented mechanisms. It was recently shown that Gly-84 residue in DP molecules prevents the binding of the invariant chain via its CLIP region on the antigen-binding groove[63]. CLIP binding is necessary to block endogenous peptide binding to class II molecules till they reach the MIIC compartment where exogenous peptide loading takes place. Therefore, DP84Gly molecules are able to present both intracellular and extracellular peptides[63]. Whether or how this contributes to immunity against HBV needs to be examined in functional studies.
The HLA locus is characterized by high gene density, high genetic diversity and extensive LD structure, limiting the ability to pinpoint the causal variants[64]. Some class II associations might not reflect primary effects given the strong LD between DQ and DR loci, and the weak LD between DQ-DR and DP loci. Hence, uncovering the LD patterns between the DP, DQ and DR alleles in the study population, rather than examining the alleles at a single locus, would help detecting the alleles with primary effects[59]. Fine mapping studies in which associated loci are interrogated with many more variants than in GWAS prove to be useful in pinpointing the causal variants and revealing potential mechanisms. An interrogation of the entire HLA locus with 5375 variants, including 4356 SNPs, 849 amino acid polymorphisms and 170 classical alleles, in 1888 Chinese case-control individuals (from a previous HBV-persistence GWAS[28]) identified four independently associated loci by stepwise conditional analysis[24]. HLA-DPβ1 amino acid polymorphisms at positions 84-87 were the strongest association, whereas HLA-C Leu-15 was the second strongest association, conferring the effect of HLA-C*07:02. The two other associated loci were novel; HLA-DRB*13 allele and rs400488. HLA-DRB*13 was associated with protection against HBV chronicity, and was in partial LD with the CFB-tagging SNP rs12614. HLA-DRB*13 allele is distinct from other DRB1 alleles by the presence of Glu-71 in pocket-4, an anchor region for epitope binding of DR molecules, implying a specific binding motif. Remarkably, HLA-DRB*13 was one of the few classical HLA alleles identified in the pre-GWAS era with highly reproducible associations across different ethnicities[65-68]. On the other hand, the biological relevance of rs400488, an eQTL for the pseudogene HLA-J, is currently unknown[24].
The effects of associated SNPs on HLA gene expression were also investigated. An eQTL analysis showed that the risk alleles of rs3077 and rs9277535 were associated with lower HLA-DPA1 and HLA-DPB1 mRNA levels, respectively, in normal liver tissues[69]. Furthermore, allelic expression imbalance assays for both SNPs confirmed that the risk alleles were expressed less abundantly than the protective alleles in liver and blood monocyte samples from heterozygous individuals[69]. Two separate genome-wide gene expression association studies previously found similar trends for the HLA-DQ SNP rs7453920 and the HLA-C SNP rs3130542. The risk variant of rs7453920 was associated with lower HLA-DQ mRNA levels in peripheral blood monocytes[53], while the risk variant of rs3130542 was associated with lower HLA-C mRNA in lymphoblastoid cell lines derived from European subjects[70]. Moreover, the susceptible HLA-C*07:02 allele, tagged by rs3130542, have an intact mir-148a binding site, and a low expression due to RNA interference[71]. These results suggested that reduced expression of HLA-DP and HLA-DQ molecules in antigen-presenting cells, and reduced expression of HLA-C molecule in hepatocytes might result in ineffective antigen presentation to T cells, leading to inability to clear HBV infection, and thus, HBV persistence. However, a discrepant result was noted for rs9277534_A/G, the most significant HLA-DPB1 SNP in European Americans and African Americans[46]. The risk allele rs9277534_G correlated with higher HLA-DPB1 surface protein levels in lymphoid cell lines and higher HLA-DPB1 mRNA levels in B lymphocytes isolated from healthy donors. This conflicting result can be reconciled with the previous findings by taking into account that rs9277534_G was also in strong LD with the most susceptible HLA-DPB1 allele (*01:01) in these populations. Thus, the risk-conferring effect of HLA-DPB1*01:01 probably prevailed over the protective effect of its increased expression.
To sum up, polymorphisms marking the classical HLA genes influence immune responses by altering the antigen-binding properties of HLA molecules and the expression levels thereof. The relative contributions of these effects to susceptibility to HBV persistence vary among populations depending on the allele frequencies of variants and LD relationships among them.
The standard 3 doses of hepatitis B vaccine (based on recombinant HBs antigen) have been implemented as part of the routine infantile vaccination program in more than 160 countries, as a result of which substantial declines in the prevalence of chronic HBV carriers and in the incidence of childhood HBV-related liver diseases have been observed[6,72]. However, there is a large natural variation in the antibody response to hepatitis B vaccine. Post-vaccination antibody titers range from undetectable levels to higher than 2000 mIU/mL[73]. Anti-HBs titers above 100 mIU/mL are referred to as successful vaccination, whereas anti-HBs titers below 10 mIU/mL as non-protective. Individuals that mount an antibody response below this threshold, i.e., non-responders, are at a high risk of HBV infection. Booster vaccination can trigger an antibody response at protective levels in a subset of non-responders[74].
Three GWAS were implemented to date to identify the genetic factors that underlie the variation in the immune response to hepatitis B vaccine (Figure 1, Table 3). The first GWAS was conducted in Indonesians by grouping vaccine recipients into three classes (i.e., low, intermediate and high) on the basis of their post-vaccination antibody titers[73]. Stepwise conditional analysis revealed three independently associated haplotype blocks within the HLA region. The first haplotype block, tagged by rs3135363, encompassed HLA-DRA and BTNL2. HLA-DRA encodes the sole α chain for HLA-DR molecules, and BTNL2 encodes a transmembrane protein involved in the negative regulation of T cell activation. BTNL2 was previously associated with autoimmune diseases[75]. The second haplotype block, tagged by rs9277535, encompassed HLA-DPA1 and HLA-DPB1 genes, suggesting that HLA-DP variants contribute to both HBV persistence and non-response to vaccine. A shared genetic basis for these traits is conceivable given that anti-HBs antibody production is a marker for both successful immune response to vaccine and clearance of HBV infection[3]. Lastly, the third haplotype block, tagged by rs9267665, was located within the class III region, consisting of many genes found in strong LD. Therefore, further research is needed to pinpoint the causal genes[73].
| Ref. | Study population | SNPID | Minor/major alleles | Riskallele | P | OR | Location | Nearest gene | Functional class |
| Png et al[73] | Indonesian (n = 3614)1 | rs3135363 | C/T | C | 6.53 × 10-22 | 1.53 | 6p21.32 | BTNL2, HLA-DRA | Intergenic |
| rs9277535 | A/G | G | 2.91 × 10-12 | 0.72 | 6p21.32 | HLA-DPB1 | 3'UTR | ||
| rs9267665 | T/C | T | 1.24 × 10-17 | 2.05 | 6p21.33 | STK19 | Intron | ||
| Pan et al[76] | Chinese (n = 1944)2 | rs477515 | T/C | T | 2.63 × 10-19 | 2.05 | 6p21.32 | HLA-DRB1 | Intergenic |
| Wu et al[81] | Taiwanese (n = 285)3 | rs7770370 | A/G | G | 1.20 × 10-08 | 0.33 | 6p21.32 | HLA-DPB1 | Intergenic |
The second GWAS for vaccine nonresponse used Chinese adults with marginal phenotypes: non-responders (< 10 mIU/mL) after booster vaccination vs high-responders (> 1000 mIU/mL) after primary vaccination[76]. The strongest associations (led by rs477515) were detected within the HLA-DR locus, and HLA-DRB1*07:01 was significantly associated with nonresponse to hepatitis B vaccination[76]. Numerous studies previously associated DRB1*07:01 with both vaccine non-response and persistent HBV infection in multiple ethnic populations[77-79]. Furthermore, DRB1*07 was also associated with decreased risk of HBV-related cirrhosis in a Turkish population[80], implicating a DRB1*07-associated hypo-immune profile against HBV.
The third GWAS used subjects with detectable and undetectable (> or < 1 mIU/mL, respectively) post-booster antibody titers from a cohort of booster vaccine recipient Taiwanese adolescents who had not responded to primary vaccination as infants[81]. Significant associations were mapped to the HLA-DP locus. The lead SNP rs7770370 was in strong LD with rs9277535. HLA-DPB1 alleles *02:01, *02:02, *03:01, *04:01 and *14:01 (encoding 84GGPM87) were associated with vaccine response, whereas *05:01 and *09:01 (encoding 84DEAV87) were associated with vaccine nonresponse[74,81]. These associations were further confirmed in a Japanese population[82]. Hence, HLA-DPB1 alleles influenced the outcome of vaccine response in the same manner as of HBV persistence, corroborating further that the ability to present the same HBs epitopes is critical in both traits. In line with this, HLA-DP SNPs (e.g., rs7770370, rs9277535 and rs3077) were also associated with vaccine nonresponse in independent replication studies in Korean infants[83] and Japanese medical students[84]; and vaccine nonresponse-associated rs477515 (HLA-DRB1) also correlated with HBV persistence in a Chinese population[30]. However, HLA-DQ SNPs (rs2856718 and rs7453920) were not associated with vaccine responsiveness[84]. Therefore, the genetic bases of HBV persistence and nonresponse to hepatitis B vaccine overlap considerably, but not completely.
Host genetic variation underlying the clinical heterogeneity of chronic HBV infections has been the topic of GWAS, too (Figure 1, Table 4). A GWAS used 648 HBV-infected Saudi Arabian subjects, comprising 343 inactive carriers, 249 patients with CHB, and 76 patients with end-stage liver diseases (i.e., cirrhosis and HCC), to study liver disease progression in chronic HBV infection[85]. The strongest association, rs2724432, was obtained using cases with end-stage liver diseases and controls with inactive HBV infection. rs2724432 was located upstream of FDX1, the product of which is involved in electron transfer from NADH to cytochrome P450. FDX1 plays a role in steroid and vitamin D synthesis in the adrenals, and bile acid synthesis in the liver[85]. Further functional assays are necessary to confirm the association of rs2724432 and the underlying molecular mechanisms. Some of the GWAS for HBV persistence contained in their case groups chronic HBV carriers with and without end-stage liver diseases. At least one GWAS employed a secondary genome-wide analysis using these sub-groups[25]; however, failed to identify any significant association, possibly due to small sample size.
| Ref. | Study population | Participant phenotypes | SNPID | Minor/major alleles | Risk allele | P | OR | Location | Nearestgene | Functional class |
| Al-Qahtani et al[85] | Saudi Arabian (n = 693) | LC/HCC vs Inactive1 | rs2724432 | T/C | T | 4.29 × 10-08 | 3.01 | 11q22.3 | FDX1 | Intergenic |
| Zhang et al[88] | Chinese (n = 4107) | HCC vs Non-HCC2 | rs17401966 | G/A | A | 3.40 × 10-19 | 0.62 | 1p36.22 | KIF1B | Intron |
| Chan et al[91] | Chinese (n = 1420) | HCC vs Non-HCC2 | rs12682266 | A/G | G | 3.76 × 10-05 | 1.38 | 8p12 | Expressed sequenced tag | Intergenic |
| Li et al[89] | Chinese (n = 12159) | HCC vs Non-HCC2 | rs9272105 | A/G | A | 5.24 × 10-22 | 1.28 | 6p21.32 | HLA-DQA1, HLA-DRB1 | Intergenic |
| rs455804 | A/C | C | 5.24 × 10-10 | 0.84 | 21q21.3 | GRIK1 | Intron | |||
| Jiang et al[90] | Chinese (n = 11799) | HCC vs Non-HCC2 | rs7574865 | T/G | G | 2.48 × 10-10 | 1.21 | 2q32.2-2q32.3 | STAT4 | Intron |
| rs9275319 | G/A | A | 2.72 × 10-17 | 1.49 | 6p21.32 | HLA-DQB1, HLA-DQA2 | Intergenic | |||
| Tan et al[86] | Chinese (n = 3387) | ACLF vs Inactive3 | rs3129859 | C/G | C | 2.64 × 10-20 | 1.83 | 6p21.32 | HLA-DQB1, HLA-DRA | Intergenic |
A GWAS in Han Chinese investigated the risk loci for acute-on-chronic liver failure (ACLF) in chronic HBV carriers, a life-threatening condition that develops as a result of sudden acute exacerbation of chronic infection[86]. rs3129859 within the HLA-DR locus was associated with HBV-related ACLF. Subsequent in silico genotyping of HLA alleles and conditional association analysis showed that the effect of rs3129859 was dependent on the HLA-DRB1*12:02 allele[86]. Concordant with this finding, HLA-DRB1*12 was previously associated with the development of HBV-related cirrhosis and HCC in Han Chinese[87].
To find the genetic risk loci for HBV-related HCC, several GWAS were implemented using HBV carriers with HCC as cases and HCC-free HBV carriers as controls[88-92] (Table 4). All of these GWAS used Chinese cohorts, exploiting the facts that more than half of HCC cases were found in China, and 80% of HCC incidences were attributable to HBV[93]. The first GWAS identified rs17401966, located in KIF1B on chromosome 1[88]. KIF1B encodes a kinesin protein involved in organelle and vesicle transport, and is a putative tumor suppressor[94]. The protective variant of rs17401966 significantly correlated with higher KIF1B protein levels in tumor-adjacent tissues, but not in HCC tissues[88]. The haplotype block tagged by rs17401966 also encompassed the entire PGD gene and the 3’ end of UBE4B gene. The products of these genes are involved in pentose phosphate metabolism and multiubiquitin chain assembly, respectively. Additional studies are necessary to identify the causal gene in this locus. Another GWAS identified rs9272105 within the HLA region and rs455804 on chromosome 21[89]. rs9272105 was located in the intergenic region between HLA-DQA1 and HLA-DRB1. HLA-DRB1 alleles *04:05 and *09:01 only partially accounted for the association of rs9272105, implying additional risk variants in LD with this SNP. rs455804 was located in intron 1 of GRIK1, which encodes an ionotropic glutamate receptor, suggesting involvement of glutamate signaling in hepatocarcinogenesis. Two additional risk loci were revealed in another GWAS: rs7574865 and rs9275319[90]. rs7574865 tagged STAT4 on chromosome 2. STAT4 is a transcription factor with important roles in the regulation of antiviral immune responses; it induces IFN-γ production in response to stimulation by interleukin-12 and type I interferons (IFN-α and IFN-β)[95]. HCC risk-associated variant of rs7574865 correlated with lower STAT4 mRNA expression in both HCC and non-tumor tissues[90]. rs9275319 tagged HLA-DQB1 and HLA-DQA2, and its effect on HCC risk was only partially accounted for by DQB1*04:01 and DQA1*03:03 alleles, indicating other risk variants in the associated locus.
SNPs identified in GWAS for HBV-related HCC were further investigated in replication studies. Intriguingly, no evidence was found for the association of rs17401966 (KIF1B) with HCC risk in chronic HBV carriers in subsequent GWAS and other replication studies in Chinese[36,96], Japanese[97], Korean[97], and Thai populations[98]. Moreover, rs17401966 was also not associated with HBV infection persistence in Chinese[99], Japanese[23] and Saudi Arabian populations[100]. Comparable allele frequencies were detected between different subgroups of chronic carriers (namely, inactive carriers, active carriers, cirrhosis and HCC) in Saudi Arabian and Chinese populations[100,101]. These results indicated that rs17401966 was a risk factor neither for HBV infection persistence nor for advancement of liver disease.
Replication studies for rs7574865 (STAT4) produced inconsistent results. The association of rs7574865 with HBV-related HCC was validated in Vietnamese[102] and Korean[103] populations, but not in two separate Chinese populations[38,96]. The effect of rs7574865_T/G on susceptibility to persistent HBV infection was also examined. HCC risk allele rs7574865_G was significantly associated with chronic HBV infection in Koreans[103] and Chinese[104]. However, this association was not reproduced in another Chinese population[38]. Lastly, rs7574865_G was also significantly associated with the risk of HBV-related cirrhosis in a Chinese population[101]. Overall, these results supported a role for STAT4 polymorphisms in HBV infection outcomes in a population specific manner.
The associations of the two HBV-related HCC risk SNPs within the HLA-DQ/DR locus (rs9275319 and rs9272105) were validated by an independent replication study in Chinese subjects[105], whereas two separate studies in Koreans[103] and in Chinese[106] failed to replicate the association of rs9275319. rs9275319 was also associated with susceptibility to HBV persistence in the original GWAS[90], and in other replication studies performed in Korean[103] and Chinese populations[30,106]. Moreover, rs9275319 was associated with increased risk of cirrhosis in Chinese HBV carriers[101]. Therefore, it might be speculated that rs9275319 was associated with an ineffective immune profile against HBV that is too weak to clear HBV infection, yet strong enough to maintain a state of basal liver necroinflammation leading to advanced liver diseases. On the other hand, the effect of rs9272105 on HBV persistence is not yet clear. HCC risk-associated variant of rs9272105 conferred susceptibility to chronic HBV infection in one Chinese population[30], and protection against chronic HBV infection in another Chinese population[89]. These conflicting results indicated that rs9272105 did not have a primary effect on HBV persistence.
To fully explore whether the genetic bases of HBV persistence and HBV-related liver diseases overlap, SNPs identified in GWAS for HBV persistence were also examined for possible effects on the development of advanced liver diseases. Most studies showed that HLA-DP variants (rs3077 and rs9277535) were associated neither with progression from inactive carrier state to disease-active states[35,37,39,45], nor with development of end-stage liver diseases[31,35,38,39,47]. Similar results were found for HLA-DQ variants (rs2856718 and rs7453920) regarding the development of end-stage liver diseases[38,39,47]. Only few studies reported significant associations of these SNPs with HBV-related HCC risk[36,89,107]. A meta-analysis with 4864 HBV-positive HCC cases and 29790 HBV-infected controls also failed to associate rs3077 and rs9277535 with HCC risk[44]. Lastly, none of the 13 HBV persistence risk SNPs that were identified hitherto in GWAS were found to be associated with HBV-related HCC in a population of 1161 cases and 1353 controls[29]. Therefore, SNPs that were associated with HBV persistence/clearance had minimal, if any, effects on progression of liver disease in chronic carriers. These results implied that distinct immune mechanisms might be critical at different stages of HBV infection, probably reflecting the evolving dynamics between HBV and the host immune system during chronic infections.
It is important to have a comprehensive understanding of the mechanisms of failure to clear HBV infection, and translate this knowledge into effective therapies to prevent the development of chronicity and other adverse outcomes. With this motive, many GWAS were performed to dissect the genetic basis of HBV-related traits. These GWAS provided novel insights into the pathogenesis of HBV persistence and non-response to hepatitis B vaccine. For instance, the association of HLA-DPB1 with HBV persistence was unknown before GWAS. DPB1 gene was neglected in previous candidate gene-based association studies, mainly because it was less polymorphic than DQB1 and DRB genes[46]. In addition to the classical HLA genes HLA-DP, HLA-DQ and HLA-C, GWAS for HBV persistence provided candidate genes including non-classical HLA genes (EHMT2, TCF19, CFB, NOTCH4) and non-HLA genes (UBE2L3, CD40, INTS10), indicating that genetic susceptibility to HBV persistence is determined by regulation of immune responses at multiple levels. Many of these candidate genes had known roles in immune responses, whereas some were novel. Besides improving our understanding of the molecular mechanisms underlying HBV-related pathologies, GWAS findings are likely to have other immediate and long-running clinical implications. For instance, genetic markers provided by GWAS can be used to predict the individuals that are at a higher risk for worse prognosis, demanding a closer medical surveillance. Furthermore, identification of novel protective HLA allotypes might be exploited for the development of more effective vaccines based on alternative epitopes.
Despite their initial success, some of these GWAS also had certain limitations, including small sample sizes, lack of information on age at first infection, transmission route, maternal status and HBV genotype, unknown infection history of controls, unknown infection status for HCV and HIV, and imperfect match of age, gender and genetic background among some case-control groups. GWAS for HBV-related advanced liver diseases additionally suffered from the stochasticity of disease pathogenesis. For instance, the randomness of HBV DNA integration events in the host genome adds to the heterogeneity of HBV-related HCC[108]. Cirrhosis and HCC typically occur more than 20 years after the initial infection[109]. Smoking, alcohol consumption, aflatoxin exposure, obesity, diabetes, and treatment interventions during this period are strong confounders that also lead to clinical heterogeneity[110]. The lack of consistent associations in GWAS for HBV-related advanced liver diseases might be due to these confounders. Hence, these GWAS must be designed in a better way to account for each of these factors[111,112].
Interestingly, the susceptible alleles in the HLA-DP/DQ locus are more frequent in Asian populations than in Caucasians, which might be one of the causes for higher prevalence of chronic HBV infections in Asia[22,113]. However, most of these susceptible alleles are not frequent in Africa, too, even though chronic hepatitis B is as prevalent in Africa as in Asia. Given that the predominant mode of transmission and HBV genotypes are different in Asia and Africa (vertical vs horizontal; B, C vs A, D, E; respectively), the genetic architecture for predisposition to chronic HBV infection also varies among these populations[7,113]. Hence, allele frequency distributions across human populations might give important insights into human-HBV co-evolution.
GWAS for HBV-related traits have almost exclusively been implemented in Asian populations. So far, there is no GWAS in Africans, and only one GWAS in Saudi Arabians, where the dominant HBV genotype is D[7]. Populations with different ancestries harbor different haplotype structures and allele frequencies; therefore, implementing GWAS in other populations might also help narrow down the associated loci. This is especially desirable for fine mapping studies. Fine mapping studies on the HLA locus, where extensive LD structure hampers the detection of causal variants, are of utmost importance since immune-related genes are concentrated in this region. The findings of such studies would reveal novel insights into the pathogenesis of HBV-related traits. Therefore, additional GWAS and fine mapping studies, implemented with more refined case-control designs, larger samples, and in other ethnic populations, would further improve our understanding of HBV infection pathophysiology.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: Turkey
Peer-review report classification
Grade A (Excellent): A
Grade B (Very good): B, B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Aghakhani A, Enomoto M, Gong ZG, Tai DI S- Editor: Wang XJ L- Editor: A E- Editor: Yin SY
| 1. | Schweitzer A, Horn J, Mikolajczyk RT, Krause G, Ott JJ. Estimations of worldwide prevalence of chronic hepatitis B virus infection: a systematic review of data published between 1965 and 2013. Lancet. 2015;386:1546-1555. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1806] [Cited by in F6Publishing: 1913] [Article Influence: 212.6] [Reference Citation Analysis (3)] |
| 2. | Lozano R, Naghavi M, Foreman K, Lim S, Shibuya K, Aboyans V, Abraham J, Adair T, Aggarwal R, Ahn SY. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380:2095-2128. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9500] [Cited by in F6Publishing: 9328] [Article Influence: 777.3] [Reference Citation Analysis (0)] |
| 3. | Trépo C, Chan HL, Lok A. Hepatitis B virus infection. Lancet. 2014;384:2053-2063. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1004] [Cited by in F6Publishing: 1085] [Article Influence: 108.5] [Reference Citation Analysis (0)] |
| 4. | Beasley RP, Hwang LY, Lin CC, Chien CS. Hepatocellular carcinoma and hepatitis B virus. A prospective study of 22 707 men in Taiwan. Lancet. 1981;2:1129-1133. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1838] [Cited by in F6Publishing: 1730] [Article Influence: 40.2] [Reference Citation Analysis (0)] |
| 5. | Perz JF, Armstrong GL, Farrington LA, Hutin YJ, Bell BP. The contributions of hepatitis B virus and hepatitis C virus infections to cirrhosis and primary liver cancer worldwide. J Hepatol. 2006;45:529-538. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1764] [Cited by in F6Publishing: 1798] [Article Influence: 99.9] [Reference Citation Analysis (0)] |
| 6. | World Health Organization. Global Hepatitis Report 2017. Accessed October 2017. [ISBN: 978-92-4-156545-5]. Available from: http://www.who.int/hepatitis/publications/global-hepatitis-report2017/en/. [Cited in This Article: ] |
| 7. | McMahon BJ. The natural history of chronic hepatitis B virus infection. Hepatology. 2009;49:S45-S55. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 519] [Cited by in F6Publishing: 537] [Article Influence: 35.8] [Reference Citation Analysis (0)] |
| 8. | Liang TJ. Hepatitis B: the virus and disease. Hepatology. 2009;49:S13-S21. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 554] [Cited by in F6Publishing: 591] [Article Influence: 39.4] [Reference Citation Analysis (0)] |
| 9. | Hoofnagle JH, Doo E, Liang TJ, Fleischer R, Lok AS. Management of hepatitis B: summary of a clinical research workshop. Hepatology. 2007;45:1056-1075. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 438] [Cited by in F6Publishing: 420] [Article Influence: 24.7] [Reference Citation Analysis (0)] |
| 10. | Nguyen VT, Law MG, Dore GJ. Hepatitis B-related hepatocellular carcinoma: epidemiological characteristics and disease burden. J Viral Hepat. 2009;16:453-463. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 205] [Cited by in F6Publishing: 221] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 11. | He YL, Zhao YR, Zhang SL, Lin SM. Host susceptibility to persistent hepatitis B virus infection. World J Gastroenterol. 2006;12:4788-4793. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 37] [Cited by in F6Publishing: 39] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 12. | Lin TM, Chen CJ, Wu MM, Yang CS, Chen JS, Lin CC, Kwang TY, Hsu ST, Lin SY, Hsu LC. Hepatitis B virus markers in Chinese twins. Anticancer Res. 1989;9:737-741. [PubMed] [Cited in This Article: ] |
| 13. | Höhler T, Reuss E, Evers N, Dietrich E, Rittner C, Freitag CM, Vollmar J, Schneider PM, Fimmers R. Differential genetic determination of immune responsiveness to hepatitis B surface antigen and to hepatitis A virus: a vaccination study in twins. Lancet. 2002;360:991-995. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 147] [Cited by in F6Publishing: 145] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 14. | Newport MJ, Goetghebuer T, Weiss HA, Whittle H, Siegrist CA, Marchant A; MRC Gambia Twin Study Group. Genetic regulation of immune responses to vaccines in early life. Genes Immun. 2004;5:122-129. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 155] [Cited by in F6Publishing: 165] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 15. | Singh R, Kaul R, Kaul A, Khan K. A comparative review of HLA associations with hepatitis B and C viral infections across global populations. World J Gastroenterol. 2007;13:1770-1787. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 161] [Cited by in F6Publishing: 168] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 16. | Wang L, Zou ZQ, Wang K. Clinical Relevance of HLA Gene Variants in HBV Infection. J Immunol Res. 2016;2016:9069375. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 43] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 17. | Matsuura K, Isogawa M, Tanaka Y. Host genetic variants influencing the clinical course of hepatitis B virus infection. J Med Virol. 2016;88:371-379. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 41] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 18. | Tong Hv, Bock CT, Velavan TP. Genetic insights on host and hepatitis B virus in liver diseases. Mutat Res Rev Mutat Res. 2014;762:65-75. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 11] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 19. | Katrinli S, Nigdelioglu A, Ozdil K, Dinler-Doganay G, Doganay L. The association of variations in TLR genes and spontaneous immune control of hepatitis B virus. Clin Res Hepatol Gastroenterol. 2018;42:139-144. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 5] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 20. | Katrinli S, Enc FY, Ozdil K, Ozturk O, Tuncer I, Doganay GD, Doganay L. Effect of HLA-DPA1 alleles on chronic hepatitis B prognosis and treatment response. North Clin Istanb. 2017;3:168-174. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1] [Cited by in F6Publishing: 1] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 21. | Doganay L, Tuncer I, Katrinli S, Enc FY, Ozturk O, Colak Y, Ulasoglu C, Dinler G. The effect of HLA-DQB1 alleles on virologic breakthroughs during chronic hepatitis B treatment with genetically low barrier drugs. Clin Res Hepatol Gastroenterol. 2013;37:359-364. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in F6Publishing: 5] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 22. | Kamatani Y, Wattanapokayakit S, Ochi H, Kawaguchi T, Takahashi A, Hosono N, Kubo M, Tsunoda T, Kamatani N, Kumada H. A genome-wide association study identifies variants in the HLA-DP locus associated with chronic hepatitis B in Asians. Nat Genet. 2009;41:591-595. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 396] [Cited by in F6Publishing: 407] [Article Influence: 27.1] [Reference Citation Analysis (0)] |
| 23. | Mbarek H, Ochi H, Urabe Y, Kumar V, Kubo M, Hosono N, Takahashi A, Kamatani Y, Miki D, Abe H. A genome-wide association study of chronic hepatitis B identified novel risk locus in a Japanese population. Hum Mol Genet. 2011;20:3884-3892. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 165] [Cited by in F6Publishing: 178] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 24. | Zhu M, Dai J, Wang C, Wang Y, Qin N, Ma H, Song C, Zhai X, Yang Y, Liu J. Fine mapping the MHC region identified four independent variants modifying susceptibility to chronic hepatitis B in Han Chinese. Hum Mol Genet. 2016;25:1225-1232. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 27] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 25. | Nishida N, Sawai H, Matsuura K, Sugiyama M, Ahn SH, Park JY, Hige S, Kang JH, Suzuki K, Kurosaki M. Genome-wide association study confirming association of HLA-DP with protection against chronic hepatitis B and viral clearance in Japanese and Korean. PLoS One. 2012;7:e39175. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 109] [Cited by in F6Publishing: 123] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 26. | Kim YJ, Kim HY, Lee JH, Yu SJ, Yoon JH, Lee HS, Kim CY, Cheong JY, Cho SW, Park NH. A genome-wide association study identified new variants associated with the risk of chronic hepatitis B. Hum Mol Genet. 2013;22:4233-4238. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 85] [Cited by in F6Publishing: 92] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 27. | Chang SW, Fann CS, Su WH, Wang YC, Weng CC, Yu CJ, Hsu CL, Hsieh AR, Chien RN, Chu CM. A genome-wide association study on chronic HBV infection and its clinical progression in male Han-Taiwanese. PLoS One. 2014;9:e99724. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 44] [Cited by in F6Publishing: 43] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 28. | Hu Z, Liu Y, Zhai X, Dai J, Jin G, Wang L, Zhu L, Yang Y, Liu J, Chu M. New loci associated with chronic hepatitis B virus infection in Han Chinese. Nat Genet. 2013;45:1499-1503. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 111] [Cited by in F6Publishing: 118] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 29. | Jiang DK, Ma XP, Yu H, Cao G, Ding DL, Chen H, Huang HX, Gao YZ, Wu XP, Long XD. Genetic variants in five novel loci including CFB and CD40 predispose to chronic hepatitis B. Hepatology. 2015;62:118-128. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 62] [Cited by in F6Publishing: 64] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 30. | Li Y, Si L, Zhai Y, Hu Y, Hu Z, Bei JX, Xie B, Ren Q, Cao P, Yang F. Genome-wide association study identifies 8p21.3 associated with persistent hepatitis B virus infection among Chinese. Nat Commun. 2016;7:11664. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 42] [Cited by in F6Publishing: 45] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 31. | An P, Winkler C, Guan L, O’Brien SJ, Zeng Z; HBV Study Consortium. A common HLA-DPA1 variant is a major determinant of hepatitis B virus clearance in Han Chinese. J Infect Dis. 2011;203:943-947. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 48] [Cited by in F6Publishing: 54] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 32. | Lau KC, Lam CW, Law CY, Lai ST, Tsang TY, Siu CW, To WK, Leung KF, Mak CM, Poon WT. Non-invasive screening of HLA-DPA1 and HLA-DPB1 alleles for persistent hepatitis B virus infection: susceptibility for vertical transmission and toward a personalized approach for vaccination and treatment. Clin Chim Acta. 2011;412:952-957. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 14] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 33. | Wang L, Wu XP, Zhang W, Zhu DH, Wang Y, Li YP, Tian Y, Li RC, Li Z, Zhu X. Evaluation of genetic susceptibility loci for chronic hepatitis B in Chinese: two independent case-control studies. PLoS One. 2011;6:e17608. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 56] [Cited by in F6Publishing: 60] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 34. | Guo X, Zhang Y, Li J, Ma J, Wei Z, Tan W, O’Brien SJ. Strong influence of human leukocyte antigen (HLA)-DP gene variants on development of persistent chronic hepatitis B virus carriers in the Han Chinese population. Hepatology. 2011;53:422-428. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 101] [Cited by in F6Publishing: 114] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 35. | Li J, Yang D, He Y, Wang M, Wen Z, Liu L, Yao J, Matsuda K, Nakamura Y, Yu J. Associations of HLA-DP variants with hepatitis B virus infection in southern and northern Han Chinese populations: a multicenter case-control study. PLoS One. 2011;6:e24221. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 43] [Cited by in F6Publishing: 49] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 36. | Hu L, Zhai X, Liu J, Chu M, Pan S, Jiang J, Zhang Y, Wang H, Chen J, Shen H. Genetic variants in human leukocyte antigen/DP-DQ influence both hepatitis B virus clearance and hepatocellular carcinoma development. Hepatology. 2012;55:1426-1431. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 127] [Cited by in F6Publishing: 139] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 37. | Wong DK, Watanabe T, Tanaka Y, Seto WK, Lee CK, Fung J, Lin CK, Huang FY, Lai CL, Yuen MF. Role of HLA-DP polymorphisms on chronicity and disease activity of hepatitis B infection in Southern Chinese. PLoS One. 2013;8:e66920. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 33] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 38. | Liao Y, Cai B, Li Y, Chen J, Tao C, Huang H, Wang L. Association of HLA-DP/DQ and STAT4 polymorphisms with HBV infection outcomes and a mini meta-analysis. PLoS One. 2014;9:e111677. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25] [Cited by in F6Publishing: 26] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 39. | Tao J, Su K, Yu C, Liu X, Wu W, Xu W, Jiang B, Luo R, Yao J, Zhou J. Fine mapping analysis of HLA-DP/DQ gene clusters on chromosome 6 reveals multiple susceptibility loci for HBV infection. Amino Acids. 2015;47:2623-2634. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 9] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 40. | Zhang X, Jia J, Dong J, Yu F, Ma N, Li M, Liu X, Liu W, Li T, Liu D. HLA-DQ polymorphisms with HBV infection: different outcomes upon infection and prognosis to lamivudine therapy. J Viral Hepat. 2014;21:491-498. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 22] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 41. | Posuwan N, Payungporn S, Tangkijvanich P, Ogawa S, Murakami S, Iijima S, Matsuura K, Shinkai N, Watanabe T, Poovorawan Y. Genetic association of human leukocyte antigens with chronicity or resolution of hepatitis B infection in thai population. PLoS One. 2014;9:e86007. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 25] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 42. | Wasityastuti W, Yano Y, Ratnasari N, Triyono T, Triwikatmani C, Indrarti F, Heriyanto DS, Yamani LN, Liang Y, Utsumi T. Protective effects of HLA-DPA1/DPB1 variants against Hepatitis B virus infection in an Indonesian population. Infect Genet Evol. 2016;41:177-184. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 17] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 43. | Liao Y, Cai B, Li Y, Chen J, Ying B, Tao C, Zhao M, Ba Z, Zhang Z, Wang L. Association of HLA-DP/DQ, STAT4 and IL-28B variants with HBV viral clearance in Tibetans and Uygurs in China. Liver Int. 2015;35:886-896. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 22] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 44. | Yu L, Cheng YJ, Cheng ML, Yao YM, Zhang Q, Zhao XK, Liu HJ, Hu YX, Mu M, Wang B. Quantitative assessment of common genetic variations in HLA-DP with hepatitis B virus infection, clearance and hepatocellular carcinoma development. Sci Rep. 2015;5:14933. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 19] [Cited by in F6Publishing: 21] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 45. | Vermehren J, Lötsch J, Susser S, Wicker S, Berger A, Zeuzem S, Sarrazin C, Doehring A. A common HLA-DPA1 variant is associated with hepatitis B virus infection but fails to distinguish active from inactive Caucasian carriers. PLoS One. 2012;7:e32605. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 38] [Cited by in F6Publishing: 39] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 46. | Thomas R, Thio CL, Apps R, Qi Y, Gao X, Marti D, Stein JL, Soderberg KA, Moody MA, Goedert JJ. A novel variant marking HLA-DP expression levels predicts recovery from hepatitis B virus infection. J Virol. 2012;86:6979-6985. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 132] [Cited by in F6Publishing: 123] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 47. | Al-Qahtani AA, Al-Anazi MR, Abdo AA, Sanai FM, Al-Hamoudi W, Alswat KA, Al-Ashgar HI, Khalaf NZ, Eldali AM, Viswan NA. Association between HLA variations and chronic hepatitis B virus infection in Saudi Arabian patients. PLoS One. 2014;9:e80445. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 43] [Cited by in F6Publishing: 45] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 48. | Trinks J, Nishida N, Hulaniuk ML, Caputo M, Tsuchiura T, Marciano S, Haddad L, Blejer J, Bartoli S, Ameigeiras B. Role of HLA-DP and HLA-DQ on the clearance of hepatitis B virus and the risk of chronic infection in a multiethnic population. Liver Int. 2017;37:1476-1487. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 16] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 49. | Scheer S, Zaph C. The Lysine Methyltransferase G9a in Immune Cell Differentiation and Function. Front Immunol. 2017;8:429. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 44] [Cited by in F6Publishing: 56] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 50. | Krautkramer KA, Linnemann AK, Fontaine DA, Whillock AL, Harris TW, Schleis GJ, Truchan NA, Marty-Santos L, Lavine JA, Cleaver O. Tcf19 is a novel islet factor necessary for proliferation and survival in the INS-1 β-cell line. Am J Physiol Endocrinol Metab. 2013;305:E600-E610. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 28] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 51. | Ferreira MA, Hottenga JJ, Warrington NM, Medland SE, Willemsen G, Lawrence RW, Gordon S, de Geus EJ, Henders AK, Smit JH. Sequence variants in three loci influence monocyte counts and erythrocyte volume. Am J Hum Genet. 2009;85:745-749. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 64] [Cited by in F6Publishing: 64] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 52. | Lewis MJ, Vyse S, Shields AM, Boeltz S, Gordon PA, Spector TD, Lehner PJ, Walczak H, Vyse TJ. UBE2L3 polymorphism amplifies NF-κB activation and promotes plasma cell development, linking linear ubiquitination to multiple autoimmune diseases. Am J Hum Genet. 2015;96:221-234. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 71] [Cited by in F6Publishing: 71] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 53. | Zeller T, Wild P, Szymczak S, Rotival M, Schillert A, Castagne R, Maouche S, Germain M, Lackner K, Rossmann H. Genetics and beyond--the transcriptome of human monocytes and disease susceptibility. PLoS One. 2010;5:e10693. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 483] [Cited by in F6Publishing: 504] [Article Influence: 36.0] [Reference Citation Analysis (0)] |
| 54. | Stoermer KA, Morrison TE. Complement and viral pathogenesis. Virology. 2011;411:362-373. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 179] [Cited by in F6Publishing: 184] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 55. | Radtke F, Fasnacht N, Macdonald HR. Notch signaling in the immune system. Immunity. 2010;32:14-27. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 371] [Cited by in F6Publishing: 412] [Article Influence: 29.4] [Reference Citation Analysis (0)] |
| 56. | Jacobson EM, Concepcion E, Oashi T, Tomer Y. A Graves’ disease-associated Kozak sequence single-nucleotide polymorphism enhances the efficiency of CD40 gene translation: a case for translational pathophysiology. Endocrinology. 2005;146:2684-2691. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 161] [Cited by in F6Publishing: 162] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 57. | Elgueta R, Benson MJ, de Vries VC, Wasiuk A, Guo Y, Noelle RJ. Molecular mechanism and function of CD40/CD40L engagement in the immune system. Immunol Rev. 2009;229:152-172. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 836] [Cited by in F6Publishing: 1033] [Article Influence: 68.9] [Reference Citation Analysis (0)] |
| 58. | Nishida N, Sawai H, Kashiwase K, Minami M, Sugiyama M, Seto WK, Yuen MF, Posuwan N, Poovorawan Y, Ahn SH. New susceptibility and resistance HLA-DP alleles to HBV-related diseases identified by a trans-ethnic association study in Asia. PLoS One. 2014;9:e86449. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 30] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 59. | Nishida N, Ohashi J, Khor SS, Sugiyama M, Tsuchiura T, Sawai H, Hino K, Honda M, Kaneko S, Yatsuhashi H. Understanding of HLA-conferred susceptibility to chronic hepatitis B infection requires HLA genotyping-based association analysis. Sci Rep. 2016;6:24767. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 37] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 60. | Díaz G, Amicosante M, Jaraquemada D, Butler RH, Guillén MV, Sánchez M, Nombela C, Arroyo J. Functional analysis of HLA-DP polymorphism: a crucial role for DPbeta residues 9, 11, 35, 55, 56, 69 and 84-87 in T cell allorecognition and peptide binding. Int Immunol. 2003;15:565-576. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 86] [Cited by in F6Publishing: 88] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 61. | Sidney J, Steen A, Moore C, Ngo S, Chung J, Peters B, Sette A. Five HLA-DP molecules frequently expressed in the worldwide human population share a common HLA supertypic binding specificity. J Immunol. 2010;184:2492-2503. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 84] [Cited by in F6Publishing: 83] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 62. | Andreatta M, Nielsen M. Characterizing the binding motifs of 11 common human HLA-DP and HLA-DQ molecules using NNAlign. Immunology. 2012;136:306-311. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 26] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 63. | Yamashita Y, Anczurowski M, Nakatsugawa M, Tanaka M, Kagoya Y, Sinha A, Chamoto K, Ochi T, Guo T, Saso K. HLA-DP84Gly constitutively presents endogenous peptides generated by the class I antigen processing pathway. Nat Commun. 2017;8:15244. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in F6Publishing: 27] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 64. | de Bakker PI, Raychaudhuri S. Interrogating the major histocompatibility complex with high-throughput genomics. Hum Mol Genet. 2012;21:R29-R36. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 70] [Cited by in F6Publishing: 75] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 65. | Thursz MR, Kwiatkowski D, Allsopp CE, Greenwood BM, Thomas HC, Hill AV. Association between an MHC class II allele and clearance of hepatitis B virus in the Gambia. N Engl J Med. 1995;332:1065-1069. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 316] [Cited by in F6Publishing: 309] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 66. | Höhler T, Gerken G, Notghi A, Lubjuhn R, Taheri H, Protzer U, Löhr HF, Schneider PM, Meyer zum Büschenfelde KH, Rittner C. HLA-DRB1*1301 and *1302 protect against chronic hepatitis B. J Hepatol. 1997;26:503-507. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 113] [Cited by in F6Publishing: 123] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 67. | Ahn SH, Han KH, Park JY, Lee CK, Kang SW, Chon CY, Kim YS, Park K, Kim DK, Moon YM. Association between hepatitis B virus infection and HLA-DR type in Korea. Hepatology. 2000;31:1371-1373. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 97] [Cited by in F6Publishing: 103] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 68. | Thio CL, Thomas DL, Karacki P, Gao X, Marti D, Kaslow RA, Goedert JJ, Hilgartner M, Strathdee SA, Duggal P. Comprehensive analysis of class I and class II HLA antigens and chronic hepatitis B virus infection. J Virol. 2003;77:12083-12087. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 101] [Cited by in F6Publishing: 111] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 69. | O’Brien TR, Kohaar I, Pfeiffer RM, Maeder D, Yeager M, Schadt EE, Prokunina-Olsson L. Risk alleles for chronic hepatitis B are associated with decreased mRNA expression of HLA-DPA1 and HLA-DPB1 in normal human liver. Genes Immun. 2011;12:428-433. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 77] [Cited by in F6Publishing: 78] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 70. | Montgomery SB, Sammeth M, Gutierrez-Arcelus M, Lach RP, Ingle C, Nisbett J, Guigo R, Dermitzakis ET. Transcriptome genetics using second generation sequencing in a Caucasian population. Nature. 2010;464:773-777. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 673] [Cited by in F6Publishing: 635] [Article Influence: 45.4] [Reference Citation Analysis (0)] |
| 71. | Kaur G, Gras S, Mobbs JI, Vivian JP, Cortes A, Barber T, Kuttikkatte SB, Jensen LT, Attfield KE, Dendrou CA. Structural and regulatory diversity shape HLA-C protein expression levels. Nat Commun. 2017;8:15924. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 84] [Cited by in F6Publishing: 74] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 72. | Zanetti AR, Van Damme P, Shouval D. The global impact of vaccination against hepatitis B: a historical overview. Vaccine. 2008;26:6266-6273. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 273] [Cited by in F6Publishing: 278] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 73. | Png E, Thalamuthu A, Ong RT, Snippe H, Boland GJ, Seielstad M. A genome-wide association study of hepatitis B vaccine response in an Indonesian population reveals multiple independent risk variants in the HLA region. Hum Mol Genet. 2011;20:3893-3898. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 84] [Cited by in F6Publishing: 95] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 74. | Wu TW, Chu CC, Ho TY, Chang Liao HW, Lin SK, Lin M, Lin HH, Wang LY. Responses to booster hepatitis B vaccination are significantly correlated with genotypes of human leukocyte antigen (HLA)-DPB1 in neonatally vaccinated adolescents. Hum Genet. 2013;132:1131-1139. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 20] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 75. | Nguyen T, Liu XK, Zhang Y, Dong C. BTNL2, a butyrophilin-like molecule that functions to inhibit T cell activation. J Immunol. 2006;176:7354-7360. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 129] [Cited by in F6Publishing: 140] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 76. | Pan L, Zhang L, Zhang W, Wu X, Li Y, Yan B, Zhu X, Liu X, Yang C, Xu J. A genome-wide association study identifies polymorphisms in the HLA-DR region associated with non-response to hepatitis B vaccination in Chinese Han populations. Hum Mol Genet. 2014;23:2210-2219. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 51] [Cited by in F6Publishing: 60] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 77. | Li Y, Ni R, Song W, Shao W, Shrestha S, Ahmad S, Cunningham CK, Flynn PM, Kapogiannis BG, Wilson CM. Clear and independent associations of several HLA-DRB1 alleles with differential antibody responses to hepatitis B vaccination in youth. Hum Genet. 2009;126:685-696. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in F6Publishing: 27] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 78. | Li ZK, Nie JJ, Li J, Zhuang H. The effect of HLA on immunological response to hepatitis B vaccine in healthy people: a meta-analysis. Vaccine. 2013;31:4355-4361. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 52] [Cited by in F6Publishing: 54] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 79. | Yan ZH, Fan Y, Wang XH, Mao Q, Deng GH, Wang YM. Relationship between HLA-DR gene polymorphisms and outcomes of hepatitis B viral infections: a meta-analysis. World J Gastroenterol. 2012;18:3119-3128. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 29] [Cited by in F6Publishing: 33] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 80. | Doganay L, Fejzullahu A, Katrinli S, Yilmaz Enc F, Ozturk O, Colak Y, Ulasoglu C, Tuncer I, Dinler Doganay G. Association of human leukocyte antigen DQB1 and DRB1 alleles with chronic hepatitis B. World J Gastroenterol. 2014;20:8179-8186. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 15] [Cited by in F6Publishing: 14] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 81. | Wu TW, Chen CF, Lai SK, Lin HH, Chu CC, Wang LY. SNP rs7770370 in HLA-DPB1 loci as a major genetic determinant of response to booster hepatitis B vaccination: results of a genome-wide association study. J Gastroenterol Hepatol. 2015;30:891-899. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 16] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 82. | Sakai A, Noguchi E, Fukushima T, Tagawa M, Iwabuchi A, Kita M, Kakisaka K, Miyasaka A, Takikawa Y, Sumazaki R. Identification of amino acids in antigen-binding site of class II HLA proteins independently associated with hepatitis B vaccine response. Vaccine. 2017;35:703-710. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 22] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 83. | Roh EY, Yoon JH, In JW, Lee N, Shin S, Song EY. Association of HLA-DP variants with the responsiveness to Hepatitis B virus vaccination in Korean Infants. Vaccine. 2016;34:2602-2607. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 12] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 84. | Okada Y, Uno N, Sato S, Mori S, Sasaki D, Kaku N, Kosai K, Morinaga Y, Hasegawa H, Yanagihara K. Strong influence of human leukocyte antigen-DP variants on response to hepatitis B vaccine in a Japanese population. Vaccine. 2017;35:5662-5665. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 5] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 85. | Al-Qahtani A, Khalak HG, Alkuraya FS, Al-hamoudi W, Alswat K, Al Balwi MA, Al Abdulkareem I, Sanai FM, Abdo AA. Genome-wide association study of chronic hepatitis B virus infection reveals a novel candidate risk allele on 11q22.3. J Med Genet. 2013;50:725-732. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 23] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 86. | Tan W, Xia J, Dan Y, Li M, Lin S, Pan X, Wang H, Tang Y, Liu N, Tan S. Genome-wide association study identifies HLA-DR variants conferring risk of HBV-related acute-on-chronic liver failure. Gut. 2018;67:757-766. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 17] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 87. | Han Y, Jiang ZY, Jiao LX, Yao C, Lin QF, Ma N, Ju RQ, Yang F, Yu JH, Chen L. Association of human leukocyte antigen-DRB1 alleles with chronic hepatitis B virus infection in the Han Chinese of Northeast China. Mol Med Rep. 2012;5:1347-1351. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1] [Cited by in F6Publishing: 5] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 88. | Zhang H, Zhai Y, Hu Z, Wu C, Qian J, Jia W, Ma F, Huang W, Yu L, Yue W. Genome-wide association study identifies 1p36.22 as a new susceptibility locus for hepatocellular carcinoma in chronic hepatitis B virus carriers. Nat Genet. 2010;42:755-758. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 253] [Cited by in F6Publishing: 280] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 89. | Li S, Qian J, Yang Y, Zhao W, Dai J, Bei JX, Foo JN, McLaren PJ, Li Z, Yang J. GWAS identifies novel susceptibility loci on 6p21.32 and 21q21.3 for hepatocellular carcinoma in chronic hepatitis B virus carriers. PLoS Genet. 2012;8:e1002791. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 143] [Cited by in F6Publishing: 152] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 90. | Jiang DK, Sun J, Cao G, Liu Y, Lin D, Gao YZ, Ren WH, Long XD, Zhang H, Ma XP. Genetic variants in STAT4 and HLA-DQ genes confer risk of hepatitis B virus-related hepatocellular carcinoma. Nat Genet. 2013;45:72-75. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 198] [Cited by in F6Publishing: 231] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 91. | Chan KY, Wong CM, Kwan JS, Lee JM, Cheung KW, Yuen MF, Lai CL, Poon RT, Sham PC, Ng IO. Genome-wide association study of hepatocellular carcinoma in Southern Chinese patients with chronic hepatitis B virus infection. PLoS One. 2011;6:e28798. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 47] [Cited by in F6Publishing: 54] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 92. | Qu LS, Jin F, Guo YM, Liu TT, Xue RY, Huang XW, Xu M, Chen TY, Ni ZP, Shen XZ. Nine susceptibility loci for hepatitis B virus-related hepatocellular carcinoma identified by a pilot two-stage genome-wide association study. Oncol Lett. 2016;11:624-632. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 14] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 93. | Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69-90. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23762] [Cited by in F6Publishing: 25335] [Article Influence: 1948.8] [Reference Citation Analysis (7)] |
| 94. | Munirajan AK, Ando K, Mukai A, Takahashi M, Suenaga Y, Ohira M, Koda T, Hirota T, Ozaki T, Nakagawara A. KIF1Bbeta functions as a haploinsufficient tumor suppressor gene mapped to chromosome 1p36.2 by inducing apoptotic cell death. J Biol Chem. 2008;283:24426-24434. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 76] [Cited by in F6Publishing: 80] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 95. | Nguyen KB, Watford WT, Salomon R, Hofmann SR, Pien GC, Morinobu A, Gadina M, O’Shea JJ, Biron CA. Critical role for STAT4 activation by type 1 interferons in the interferon-gamma response to viral infection. Science. 2002;297:2063-2066. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 390] [Cited by in F6Publishing: 391] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 96. | Chen K, Shi W, Xin Z, Wang H, Zhu X, Wu X, Li Z, Li H, Liu Y. Replication of genome wide association studies on hepatocellular carcinoma susceptibility loci in a Chinese population. PLoS One. 2013;8:e77315. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 35] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 97. | Sawai H, Nishida N, Mbarek H, Matsuda K, Mawatari Y, Yamaoka M, Hige S, Kang JH, Abe K, Mochida S. No association for Chinese HBV-related hepatocellular carcinoma susceptibility SNP in other East Asian populations. BMC Med Genet. 2012;13:47. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 31] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 98. | Sopipong W, Tangkijvanich P, Payungporn S, Posuwan N, Poovorawan Y. The KIF1B (rs17401966) single nucleotide polymorphism is not associated with the development of HBV-related hepatocellular carcinoma in Thai patients. Asian Pac J Cancer Prev. 2013;14:2865-2869. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in F6Publishing: 28] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 99. | Zhong R, Tian Y, Liu L, Qiu Q, Wang Y, Rui R, Yang BF, Duan SY, Shi JX, Miao XP. HBV-related hepatocellular carcinoma susceptibility gene KIF1B is not associated with development of chronic hepatitis B. PLoS One. 2012;7:e28839. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 21] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 100. | Al-Qahtani A, Al-Anazi M, Viswan NA, Khalaf N, Abdo AA, Sanai FM, Al-Ashgar H, Al-Ahdal M. Role of single nucleotide polymorphisms of KIF1B gene in HBV-associated viral hepatitis. PLoS One. 2012;7:e45128. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 20] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 101. | Jiang DK, Ma XP, Wu X, Peng L, Yin J, Dan Y, Huang HX, Ding DL, Zhang LY, Shi Z. Genetic variations in STAT4,C2,HLA-DRB1 and HLA-DQ associated with risk of hepatitis B virus-related liver cirrhosis. Sci Rep. 2015;5:16278. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 38] [Cited by in F6Publishing: 39] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 102. | Clark A, Gerlach F, Tong Hv, Hoan NX, Song le H, Toan NL, Bock CT, Kremsner PG, Velavan TP. A trivial role of STAT4 variant in chronic hepatitis B induced hepatocellular carcinoma. Infect Genet Evol. 2013;18:257-261. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 18] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 103. | Kim LH, Cheong HS, Namgoong S, Kim JO, Kim JH, Park BL, Cho SW, Park NH, Cheong JY, Koh I. Replication of genome wide association studies on hepatocellular carcinoma susceptibility loci of STAT4 and HLA-DQ in a Korean population. Infect Genet Evol. 2015;33:72-76. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 20] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 104. | Lu Y, Zhu Y, Peng J, Wang X, Wang F, Sun Z. STAT4 genetic polymorphisms association with spontaneous clearance of hepatitis B virus infection. Immunol Res. 2015;62:146-152. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 12] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 105. | Wen J, Song C, Jiang D, Jin T, Dai J, Zhu L, An J, Liu Y, Ma S, Qin N. Hepatitis B virus genotype, mutations, human leukocyte antigen polymorphisms and their interactions in hepatocellular carcinoma: a multi-centre case-control study. Sci Rep. 2015;5:16489. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 34] [Cited by in F6Publishing: 34] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 106. | Ji X, Zhang Q, Li B, Du Y, Yin J, Liu W, Zhang H, Cao G. Impacts of human leukocyte antigen DQ genetic polymorphisms and their interactions with hepatitis B virus mutations on the risks of viral persistence, liver cirrhosis, and hepatocellular carcinoma. Infect Genet Evol. 2014;28:201-209. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 23] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 107. | Zhang X, Zheng C, Zhou ZH, Li M, Gao YT, Jin SG, Sun XH, Gao YQ. Relationship between HLA-DP gene polymorphisms and the risk of hepatocellular carcinoma: a meta-analysis. Genet Mol Res. 2015;14:15553-15563. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 10] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 108. | Hai H, Tamori A, Kawada N. Role of hepatitis B virus DNA integration in human hepatocarcinogenesis. World J Gastroenterol. 2014;20:6236-6243. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 70] [Cited by in F6Publishing: 69] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 109. | Katrinli S, Ozdil K, Sahin A, Ozturk O, Kir G, Baykal AT, Akgun E, Sarac OS, Sokmen M, Doğanay HL. Proteomic profiling of HBV infected liver biopsies with different fibrotic stages. Proteome Sci. 2017;15:7. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 18] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 110. | Casper M, Grünhage F, Lammert F. Cancer risk in chronic hepatitis B: Do genome-wide association studies hit the mark? Hepatology. 2011;53:1390-1392. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 10] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 111. | Budhu A, Wang XW. Power play: scoring our goals for liver cancer with better GWAS study design. J Hepatol. 2011;54:823-824. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 7] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 112. | Zhu H, Wu J, Shen X. Genome-wide association study: new genetic insights into HBV/HCV-related hepatocellular carcinoma genomes. Scand J Gastroenterol. 2017;52:209-215. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 15] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 113. | Tai DI, Jeng WJ, Lin CY. A global perspective on hepatitis B-related single nucleotide polymorphisms and evolution during human migration. Hepatol Commun. 2017;1:1005-1013. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in F6Publishing: 4] [Article Influence: 0.6] [Reference Citation Analysis (0)] |