Copyright
©The Author(s) 2018.
World J Gastroenterol. Mar 28, 2018; 24(12): 1299-1311
Published online Mar 28, 2018. doi: 10.3748/wjg.v24.i12.1299
Published online Mar 28, 2018. doi: 10.3748/wjg.v24.i12.1299
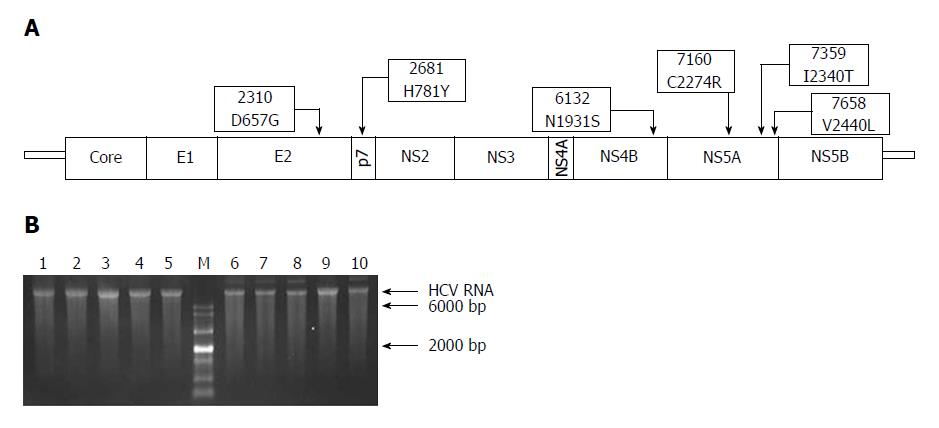
Figure 1 Schematic representation of adaptive mutations used in this study (A) and the electrophoresis results of each mutant virus RNA (B).
A: Both nucleotide substitutions (2310, 2681, 6132, 7160, 7359, and 7658) and amino acid substitutions (D657G, H781Y, N1931S, C2274R, I2340T, and V2440L) are shown; B: HCV RNA (500 ng) was analyzed using formaldehyde agarose gel electrophoresis. Lane 1: JFH1; Lane 2: JFH1-mE2; Lane 3: JFH1-mP7; Lane 4: JFH1-mNS4B; Lane 5: JFH1-mNS5A; Lane 6: JFH1-mE2/NS5A; Lane 7: JFH1-mp7/NS5A; Lane 8: JFH1-mNS4B/NS5A; Lane 9: mJFH1; Lane 10: JFH1-mE2/p7/NS5A; M: RNA marker. HCV: hepatitis C virus.
- Citation: Wang Q, Li Y, Liu SA, Xie W, Cheng J. Cell culture-adaptive mutations in hepatitis C virus promote viral production by enhancing viral replication and release. World J Gastroenterol 2018; 24(12): 1299-1311
- URL: https://www.wjgnet.com/1007-9327/full/v24/i12/1299.htm
- DOI: https://dx.doi.org/10.3748/wjg.v24.i12.1299