Copyright
©The Author(s) 2018.
World J Gastroenterol. Mar 21, 2018; 24(11): 1206-1215
Published online Mar 21, 2018. doi: 10.3748/wjg.v24.i11.1206
Published online Mar 21, 2018. doi: 10.3748/wjg.v24.i11.1206
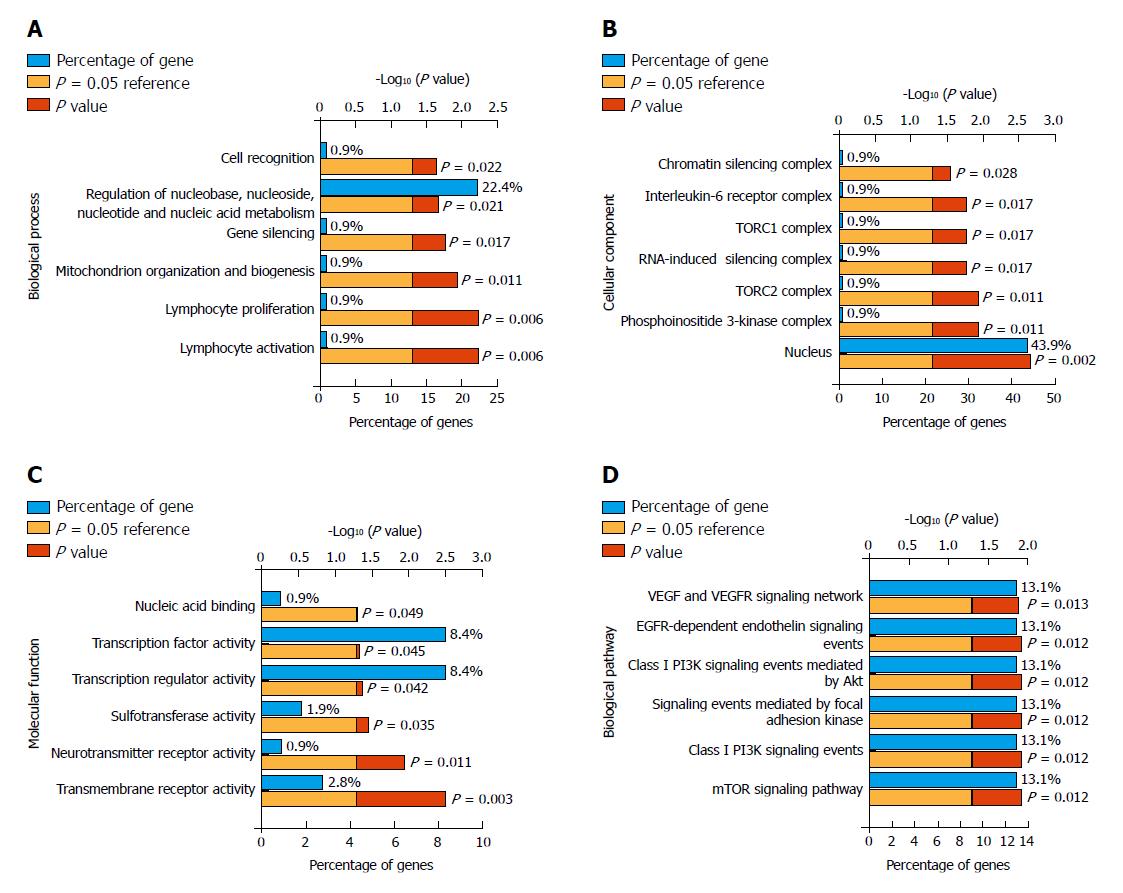
Figure 6 Genetic functional enrichment analysis.
A total of 108 consensus genes were used for functional enrichment analysis with the tool of FunRich. A: Biological process analysis showed that the genes are involved in cell recognition and gene silencing; B: Cellular component analysis indicated that the genes are portion of nucleus, chromatin silencing complex, and TORC1/2 complex; C: Molecular function results showed that the genes are involved in nucleic acid binding, transcription factor activity, and transmembrane receptor activity; D: Biological pathway analysis indicated that the genes participate in focal adhesion, PI3K, and mTOR cancer-related signaling pathways. TORC1/2: Target of rapamycin 1/2; PI3K: Phosphatidylinositide 3-kinase; mTOR: Mammalian target of rapamycin.
- Citation: Zhang C, Zhang CD, Ma MH, Dai DQ. Three-microRNA signature identified by bioinformatics analysis predicts prognosis of gastric cancer patients. World J Gastroenterol 2018; 24(11): 1206-1215
- URL: https://www.wjgnet.com/1007-9327/full/v24/i11/1206.htm
- DOI: https://dx.doi.org/10.3748/wjg.v24.i11.1206