Published online Apr 28, 2017. doi: 10.3748/wjg.v23.i16.2978
Peer-review started: December 23, 2016
First decision: January 19, 2017
Revised: February 18, 2017
Accepted: March 15, 2017
Article in press: March 15, 2017
Published online: April 28, 2017
To investigate whether hepatitis viral DNA load at 24 wk of treatment predicts response at 96 wk in patients with chronic hepatitis B.
A total of 172 hepatitis B envelope antigen (HBeAg)-positive chronic hepatitis B patients who received initial treatment at 16 tertiary hospitals in Hunan Province, China were enrolled in this study. All patients received conventional doses of lamivudine and adefovir dipivoxil, telbivudine, entecavir dispersible tablets, or entecavir tablets for 96 wk. Patients who used other antiviral drugs or antitumor and immune regulation therapy were excluded. Patients were stratified into three groups according to their viral DNA load at 24 wk: < 10 IU/mL (group 1), 10-103 IU/mL (group 2), and > 103 IU/mL (group 3). Correlations of 24-wk DNA load with HBeAg negative status and HBeAg seroconversion at 96 wk were analyzed. Receiver operating characteristic curve analysis was used to test the predictive value of the HBV DNA load at 24 wk for long-term response.
The rates of conversion to HBeAg negative status and HBeAg seroconversion rates were 53.7% and 51.9%, respectively, in group 1; 35.21% and 32.39% in group 2; and 6.38% and 6.38% in group 3. The receiver operating characteristic curves for the three subgroups revealed that the lowest DNA load (< 10 IU/mL) was better correlated with response at 96 wk than a higher DNA load (10-103 IU/mL). Nested PCR was used for amplifying and sequencing viral DNA in patients with a viral DNA load > 200 IU/mL at 96 wk; resistance mutations involving different loci were present in 26 patients, and three of these patients had a viral DNA load 10-103 IU/mL at 96 wk.
Hepatitis B viral DNA load at 24 wk of antiviral treatment in patients with chronic hepatitis B is a predictor of the viral load and response rate at 96 wk.
Core tip: Elimination of the hepatitis B surface antigen is the ultimate goal of antiviral therapy; however, this goal is rarely achieved. Complete suppression of hepatitis B virus (HBV) DNA is the current goal of antiviral therapy. Early determination of patients who are not likely to respond to chronic antiviral therapy may help providers make appropriate, timely changes. This study demonstrated a 100% complete DNA response and approximately 50% hepatitis B envelope antigen seroconversion at week 96 when the HBV DNA was suppressed to < 10 IU/mL at week 24. For patients who do not achieve HBV DNA < 10 IU/mL at week 24, add-on or alternative therapies should be considered.
- Citation: Fu XY, Tan DM, Liu CM, Gu B, Hu LH, Peng ZT, Chen B, Xie YL, Gong HY, Hu XX, Yao LH, Xu XP, Fu ZY, He LQ, Li SH, Long YZ, Li DH, Gu JL, Peng SF. Early hepatitis B viral DNA clearance predicts treatment response at week 96. World J Gastroenterol 2017; 23(16): 2978-2986
- URL: https://www.wjgnet.com/1007-9327/full/v23/i16/2978.htm
- DOI: https://dx.doi.org/10.3748/wjg.v23.i16.2978
Hepatitis B virus (HBV) infection is a pandemic disease. Although effective and safe immunization to prevent HBV infection is available, hepatitis B continues to pose a major threat to human health worldwide. An estimated 350 million or more people have had chronic HBV infection[1-3]. Those who suffer from chronic hepatitis B are at higher risk of developing cirrhosis, hepatic decompensation, and hepatocellular carcinoma (HCC). Host immune response against HBV, such as cytotoxic T lymphocyte (CTL)-associated chronic inflammation during chronic infection, can also lead to cirrhosis and liver dysfunction. It was estimated that about one million deaths due to liver failure, cirrhosis, and HCC caused by HBV infection occur every year[2].
Effective antiviral therapy can slow the progression of cirrhosis and reduce the risk of HCC in patients with chronic hepatitis B[4]. The European Association for the Study of the Liver has published clinical practice guidelines[5] for treatment of chronic hepatitis B, and an Asian-Pacific consensus statement on the management of chronic hepatitis B has been published[6]. Despite these pronouncements, universal agreement for the treatment of chronic hepatitis B has not been achieved. Orally administered nucleoside (acid) analogues strongly inhibit HBV and have a low rate of treatment discontinuation because of adverse events; thus, the drugs have been extensively used in clinical practice. Lamivudine is an antiretroviral drug that is used to prevent and treat human immunodeficiency virus (HIV) infection. It is also used to treat chronic hepatitis B. Since the United States Food and Drug Administration (FDA) approved lamivudine in 1999, adefovir, entecavir, tenofovir, and telbivudine have become available for treatment of chronic HBV infection[7-10]. Two double-blind randomized trials found that tenofovir disoproxil fumarate was superior to adefovir. Based on these results, the United States FDA approved tenofovir also for use in the treatment of chronic hepatitis B. However, the optimal course of nucleoside analogue therapy remains uncertain. Moreover, drug resistance may develop after long-term administration of nucleoside analogues. Other therapeutic strategies, such as pegylated interferon-alpha and polymerase inhibitors, have been recommended, but their effectiveness is limited to only a subset of patients. Studies on improvements of current therapies, as well as to identify early indicators that predict the long-term effects, may help to optimize the treatment effectiveness[11].
The ultimate goal of treatment of HBV infection would be functional cure, meaning a similar life expectancy of chronic HBV patients to that of patients who have self-resolution of their infection. As this clinical outcome cannot be measured in the short term, Liang et al[10] proposed apparent virological cure, which is based on the stable off-drug suppression of HBV viremia and antigenemia, and normalization of alanine aminotransaminase (ALT) and other laboratory tests. It was suggested that virological cure should be the goal of future therapies in all patients with chronic HBV infection. Hepatitis B envelope antigen (HBeAg) is an independent indicator of active viral DNA replication. It is directly correlated with disease progression to advanced stages such as cirrhosis and HCC. Therefore, sustained HBeAg seroconversion is a satisfactory result after treatment of HBeAg-positive chronic hepatitis B, and it is associated with improved long-term prognosis[5,6]. A rapid decline in hepatitis B surface antigen (HBsAg) and HBeAg titers during treatment implies a high rate of HBeAg seroconversion, as has been documented with interferon treatment[12,13]. These features are valuable in predicting the therapeutic effects in chronic HBV infection. However, very few studies using nucleoside (acid) analogue therapy have reported correlations between the changes in HBsAg and HBeAg titers and seroconversion of HBeAg[8,14].
Some authors[8,10] have proposed that patients with an undetectable HBV DNA level after the first 24 wk of antiviral therapy could have high HBeAg seroconversion rates and low drug resistance. Due to the lack of adequate data regarding these markers, no cutoff standards have been set. The sensitivity of the HBV test in patients with chronic hepatitis B has also been called into question. According to the clinical management guidelines of the European Association for the Study of the Liver Diseases[5] for chronic hepatitis B, a highly sensitive HBV DNA test should be lower than the lowest detection limit of real-time quantitative PCR (10-15 IU/mL)[5].
In this study, we aimed to further determine the response of chronic hepatitis B to antiviral treatment by using a highly sensitive HBV DNA detection kit (lower limit of detection < 10 IU/mL). Specifically, we wished to determine whether the hepatitis B viral DNA load at 24 wk of antiviral treatment is an accurate predictor of the viral load and HBeAg seroconversion rate at 96 wk, and whether our results with a more sensitive assay would differ from previously reported results.
Between December 2013 and March 2014, 172 consecutive HBeAg-positive patients who were newly diagnosed with chronic hepatitis B and received initial treatment at 16 tertiary hospitals in Hunan Province, China were enrolled in this prospective observational study. All patients were advised that they would have long-term and regular medication use and regular follow-up, and that there was the possibility of developing viral resistance and having adverse reactions to the drugs. All patients signed written informed consent forms before the start of treatment. This study was performed in accordance with the Declaration of Helsinki and approved by the Medical Ethics Committee of Xiangya Hospital, Central South University, Hunan, China. Baseline HBV DNA of all patients was ≥ 106 IU/mL, and ALT values were ≥ 2 upper limit of normal (ULN).
The following inclusion criteria were applied: patients met the diagnostic criteria for chronic hepatitis B, had no previous use of any anti-HBV drugs or other antiviral agents, had ALT values > 2 ULN, had HBV DNA > 106 IU/mL, and did not have clinically decompensated liver cirrhosis. The following patients were excluded: patients with other hepatotropic viral infections such as hepatitis C and hepatitis D, those with HIV infection, and those who used other antiviral drugs or antitumor and immune regulation therapy.
Patients were given individualized antiretroviral regimens based on the recommendations in the guideline[5] and their disease conditions and financial situations, including conventional doses of one of the following treatments for 96 wk: 100 mg/d lamivudine (LAM; GlaxoSmithKline, United Kingdom); 10 mg/d adefovir dipivoxil (ADV; Chia Tai Tianqing Pharmaceutical Group Co., Ltd., Jiangsu Province, China); 600 mg/d telbivudine (LdT, Novartis, Basel, Switzerland) once daily; 0.5 mg/d entecavir dispersible tablets (ETV; Chia Tai Tianqing Pharmaceutical Group Co., Ltd.); or 0.5 mg/d entecavir tablets (ETV; Bistrol-Myers Squibb, New York, NY). In patients who received telbivudine (33 patients), the regimen was adjusted according to the response-guided therapy to optimize the treatment as follows: if HBV DNA was greater than 300 IU/mL at 24 wk, adefovir dipivoxil was added to the regimen; this adjustment was made in 11 patients.
ALT, HBV DNA, HBsAg, HBeAg, and anti-HBe were measured in each patient before treatment and at 24, 48, 72, and 96 wk after treatment. An automated biochemical analyzer (Olympus AU640, Olympus, Japan) was used for the measurement of ALT. A chemiluminescent microparticle immunoassay (Abbott i2000, AltaVista, VA, United States) was used to detect HBsAg, HBeAg, and anti-HBe. Real-time fluorescence-based quantitative PCR was used to detect HBV DNA in a gene amplification laboratory authenticated by the Ministry of Health, China. Highly sensitive magnetic bead-based detection reagent was purchased from Hunan Shengxiang Biotechnology Co., Ltd (Northeast Gate, Hunan Province, China). The lower limit of detection with this reagent kit is 10 IU/mL, with comparable sensitivity and specificity to those with the COBAS TaqMan HBV assay for HBV DNA detection (Roche)[15,16]. A real-time PCR 7500 system was purchased from Applied Biosystems Inc. (Carlsbad, CA). The reference range of ALT was 0-40 U/L. HBsAg > 0.05 IU/mL, HBeAg > 1.0 s/co, and HBeAb < 1.0 s/co were considered positive results. Normalization of ALT was considered a biochemical response. The lowest detection limit of HBV DNA was < 10 IU/mL, and HBV DNA < 103 IU/mL was considered a complete virological response.
Detection of drug-resistance loci was carried out in each patient before the administration of antiviral therapy. At 96 wk of treatment, patients with HBV DNA > 200 IU/mL were selected for nested PCR, using their DNA as templates for detecting the presence of anti-drug mutations via PCR product sequencing. Primer sequences for amplification were A1: 5’-GCGGGGT TTTTCTTGTTGA-3’ (203-221), A2: 5’-CGGGCAACGGGGTAAAGGTTC-3’ (1158-1138), B1: 5’-CTTGTCCTCCAATTTGTCCT-3’ (345-364), and B2: 5’-ACATACTTTCCAATCAATAG-3’ (990-971). Primers A1 and A2 were used in the first round of PCR, and primers B1 and B2 were used in the second round. Reaction conditions of PCR were denaturation at 94 °C for 3 min, followed by 35 cycles of 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 60 s, and subsequently, a final extension at 72 °C for 5 min. After PCR, 5 µL PCR product from each sample was separated by 2% agarose gel electrophoresis. Amplified DNA fragment was approximately 650 bp. Positive PCR products were sequenced by Sangon Biotech (Shanghai) Co., Ltd. (Shanghai, China) using an ABI 3730xl DNA Analyzer.
SPSS 18.0 (SPSS, Chicago, IL) software was used for statistical analyses. Continuous quantitative data are presented as the means ± SD. Student’s t test was used for comparisons between groups. Chi-square (χ2) test was used for comparison of categorical data. Multivariate logistic regression was used to analyze correlations between clinical characteristics and the occurrence of conversion to HBeAg negative status/HBeAg seroconversion at 96 wk. Receiver operating characteristic (ROC) curve analysis was used to test the prediction value of viral DNA load at 24 wk for long-term response. P < 0.05 was considered statistically significant. Statistical review of the study was performed by a biomedical statistician from Public Health of Xiangya Medicine College.
A total of 243 patients were enrolled in this study, of whom 172 were followed for 96 wk, and 71 were lost to follow-up. Thus, a total of 172 patients were included in the statistical analyses. Baseline clinical data of the 172 patients are given in Table 1. Patients were divided into three groups on the basis of their HBV DNA values at 24 wk: <10 IU/mL (group 1), 10-103 IU/mL (group 2), and >103 IU/mL (group 3). No significant differences in age, ALT values, HBV, HBsAg, or HBeAg were found. The ratio of male to female patients appeared higher in groups 1 and 2 than in group 3, but the difference was not statistically significant. Moreover, we performed a correlation analysis of gender and low viral DNA load at 24 wk and found no correlation (P = 0.833).
| DNA expression level at 24-wk (IU/mL) | P value | |||
| < 10 | 10-103 | > 103 | ||
| Number of cases | 54 | 71 | 47 | |
| Gender (male/female) | 39/16 | 59/12 | 26/21 | 0.108 |
| Age (yr) | 37.79 ± 8.68 | 39.33 ± 6.92 | 35.82 ± 10.08 | 0.181 |
| ALT (U/L) median (range) | 333 (80-897) | 367 (84-813) | 319 (82-965) | 0.212 |
| AST (U/L) | 206.73 ± 133.09 | 183 ± 147.19 | 177 ± 109.85 | 0.323 |
| PLT (109/L) | 203.02 ± 70.16 | 263.33 ± 96.15 | 176.86 ± 62.03 | 0.109 |
| Total bilirubin (μmol/L) | 12.19 ± 4.07 | 11.26 ± 6.33 | 9.08 ± 100.75 | 0.791 |
| Albumin (g/L) | 43.12 ± 8.24 | 45.39 ± 7.15 | 42.29 ± 7.29 | 0.838 |
| HBV DNA (log10 IU/mL) | 7.37 ± 0.49 | 7.59 ± 0.63 | 7.26 ± 0.37 | 0.785 |
| HBsAg (log10 IU/mL) median (range) | 3.78 (2.98-5.46) | 3.66 (3.03-5.71) | 3.49 (3.04-5.66) | 0.801 |
| HBeAg (s/co) median (range) | 303.78 (1.32-5663.42) | 179.79 (1.92-6558.76) | 230.32 (1.65-6288.83) | 0.206 |
Treatment response-related variables were compared among patients grouped according to the 24-wk DNA load. As shown in Table 2, the rates of ALT normalization at 24 wk were as follows: group 1, 94.4%; group 2, 85.9%; and group 3, 40.4%. At 96 wk, the ALT normalization rates were: group 1, 100%; group 2, 93.0%; and group 3, 51.1%. Patients with HBV DNA < 103 IU/mL at 24 wk had significantly higher ALT return-to-normal rate at 96 wk than other patients (P < 0.01).
| DNA expression level at 24-wk (IU/mL) | |||
| < 10 | 10-103 | > 103 | |
| 24 wk | 94.40% | 85.9% | 40.4% |
| 96 wk | 100% | 93.0% | 51.1% |
Table 3 illustrates the correlation between the HBV DNA levels at 24 wk and the DNA response at 96 wk. Thus, 50 of the 54 (90.7%) group 1 patients had < 10 IU/mL DNA at 96 wk, and 100% were considered to have a complete response (< 103 IU/mL). In comparison, 85.9% of the group 2 patients had a complete response at 96 wk, and only 31.9% of the group 3 patients had a complete DNA response.
| 24-wk DNA (IU/mL) | DNA response at 96 wk (IU/mL) | |||
| < 10 | 10-103 | DNA complete response rate | No response (> 103) | |
| < 10 | 90.74% (50/54) | 9.26% (5/54) | 100.00% | 0 |
| 10-103 | 64.79% (46/71) | 21.13% (15/71) | 85.92% | 14.08% (10/71) |
| > 103 | 4.26% (2/47) | 27.66% (13/47) | 31.92% | 68.08% (32/47) |
Table 4 illustrates the correlation between the DNA expression at 24 wk and HBeAg conversion at 96 wk. At 96 wk, the HBeAg negative conversion and seroconversion rates in group 1 were 53.7% and 51.9%, respectively, whereas these values in group 2 were 35.2% and 32.4%, respectively. At 96 wk, only 3 of 47 patients (6.38%) in group 3 exhibited HBeAg negative conversion and seroconversion.
| HBeAg response at 96 wk | DNA expression level at 24 wk (IU/mL) | P value | ||
| < 10 | 10-103 | > 103 | ||
| Rate of conversion to HBeAg negative status | 53.70% (29/54) | 35.21% (25/71) | 6.38% (3/47) | 0.012 |
| HBeAg conversion rate | 51.85% (28/54) | 32.39% (23/71) | 6.38% (3/47) | 0.017 |
In order to determine whether the potency of nucleosides could have caused differences in viral suppression, we examined the anti-viral efficacies of different drugs. As illustrated in Table 5, of 54 patients with HBV DNA < 10 IU/mL at 24 wk, 5 (5/16 = 31.25%) received LAM + ADV, 9 (9/33 = 27.27%) received telbivudine, 21 (21/59 = 35.59%) received entecavir tablets, and 20 (20/64 = 31.25%) received entecavir dispersible tablets. No significant differences were found among different treatment groups with regard to HBV DNA below detection limits, ALT normalization rate, rate of conversion to HBeAg-negative status, and HBeAg conversion rate at 94 wk (P = 0.127).
| Virological parameter at 96 wk | LAM + ADV | Telbivudine | Entecavir tablets | Entecavir dispersible tablets | P value |
| HBV DNA below detection (< 1000 IU/mL as a reference) | 68.75% | 66.67% | 79.66% | 78.13% | 0.089 |
| ALT normalization rate | 81.25% | 75.76% | 86.44% | 85.94% | 0.096 |
| Rate of conversion to HBeAg negative status | 31.25% | 36.36% | 35.59% | 34.38% | 0.615 |
| HBeAg seroconversion rate | 25.00% | 33.33% | 32.20% | 31.25% | 0.203 |
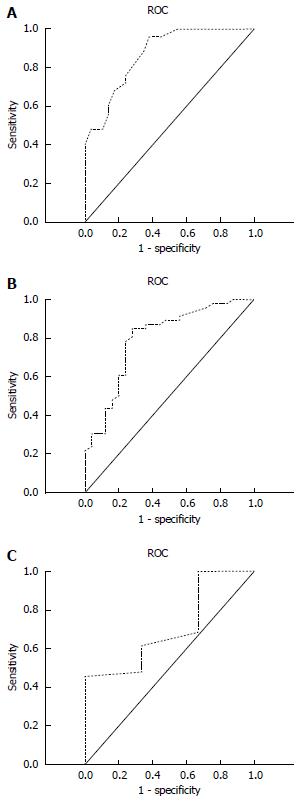
ROC curve analysis was conducted for each group to determine the predictive values of viral DNA load at 24 wk for HBeAg negative conversion or seroconversion at 96 wk (Figure 1, Table 6). The area under the curve in patients with < 10 IU/mL HBV DNA was 0.869, which was significantly larger than that in patients with 10-103 IU/mL (0.797) and in patients with > 103 IU/mL (0.505). These results suggest that the predictability of efficacy of HBV antiviral treatment at 96 wk was better among patients with < 10 IU/mL HBV DNA than among patients with 10-103 or > 103 IU/mL DNA at 24 wk.
| Group | AUC | 95%CI | Sensitivity | Specificity | PPV | NPV |
| < 10 IU/mL | 0.869 | 0.778-0.960 | 84.76% | 87.30% | 74.09% | 93.04% |
| < 1000 IU/mL | 0.797 | 0.684-0.883 | 72.00% | 82.78% | 64.18% | 87.34% |
| > 1000 IU/mL | 0.505 | 0.344-0.656 | 66.67% | 45.45% | 34.37% | 76.09% |
Patients with > 200 IU/mL HBV DNA at 96 wk underwent nested PCR for amplification and sequencing (Table 7). Various loci of drug-resistance mutations (mainly rtM204I/V, rtN236T, rtL180M, rtAl81V, and rtS202G) were present in 26 patients, of whom three had 10-103 IU/ml HBV DNA at 24 and 96 wk. Thus, in a small number of patients, persistence of modest HBVDNA levels may be the reflection of drug-resistant mutations.
| Number of cases | Resistance mutation |
| 1 | rtM204I/V + rtN236T + rtS202G |
| 1 | rtL180M + rtS202G + rtM204I/V + rtN236T |
| 1 | rtM204I/V + rtN236T |
| 1 | rtAl81V + rtM204I/V + rtN236T |
| 1 | rtM204I/V |
| 1 | rtAl81V + rtN236T + rtS202G |
| 1 | rtAl81V + rtM204I/V |
| 1 | rtL180M + rtAl81V + rtM204I/V + rtN236T |
| 1 | rtAl81V + rtM204I/V + rtS202G |
| 1 | rtAl81V + rtM204I/V + rtN236T |
| 1 | rtL180M + rtM204V |
| 2 | rtM204I + rtN236T |
| 2 | rtL180M + rtAl81V + rtM204I/V + rtN236T + rtS202G |
| 3 | rtM204I + rtN236T |
| 4 | rtN236T |
| 4 | rtAl81V + rtM204I/V + rtN236T + rtS202G |
Results in this study revealed a correlation between HBV DNA loads at 24 wk and the HBV DNA and HBeAg responses at 96 wk: Patients with a very low HBV DNA load (<10 IU/mL) at 24 wk had a 100% complete DNA response at 96 wk compared with a complete response rate in about one-third of patients who had a DNA load > 103 IU/mL at 24 wk. Similarly, HBeAg negative conversion and seroconversion rates were more favorable in patients with very low HBV DNA loads at 24 wk than in those with higher DNA loads; the conversion rate was approximately 50% at 96 wk in patients with a DNA load < 10 IU/mL, whereas it was approximately 6% in those with a DNA load > 103 IU/mL. ALT values also declined in relation to the DNA viral load at 24 wk, but the values did not change significantly between 24 and 96 wk, which may reflect decreasing hepatic inflammatory activity in the earlier stages of HBV antiviral treatment but not later. ROC curve analysis revealed that the predictability of two-year antiviral treatment efficacy was better in patients with a low initial DNA load than in patients with a higher DNA load at 24 wk. Finally, DNA sequence analysis revealed that some patients who failed to respond to anti-HBV therapy probably had drug-resistant mutations.
Based on treatment indicators at 96 wk, all the therapeutic agents used in this study (lamivudine plus ADV, telbivudine, and entecavir) appeared equally effective in treating chronic hepatitis B (Table 5).
Our findings corroborate and extend the results of few published studies on the time course of response of chronic HBV infection to nucleoside (acidic) analogue therapy. For example, after adefovir treatment for 24 wk, patients with HBV DNA fewer than 1000 copies/mL had 40% HBeAg seroconversion at 52 wk, whereas only 9% of patients whose 24-wk HBV DNA did not reach this value had HBeAg seroconversion[9,10]. Similarly, in the GLOBE study of HBeAg-positive chronic hepatitis B patients treated with telbivudine, patients who achieved complete viral suppression (< 300 copies/mL HBV DNA) at 24 wk had a HBeAg seroconversion rate of 46% at 104 wk[7]. Thus, HBeAg seroconversion was approximately 50% with HBV DNA detection limit of < 300 or < 1000 copies/mL. In this study, we used a more sensitive PCR HBV DNA assay, with the lowest limit of detection of 10 IU/mL, and found that patients with a very low HBV DNA load (<10 IU/mL) at 24 wk had a 100% complete DNA response. Further long-term studies are needed to determine whether these patients will have longer sustained undetectable levels of HBV DNA and HBeAg seroconversion. Such studies should attempt to determine whether viral replication, drug resistance, and risk of recurrence occur as long as detectable HBV DNA remains present
Functional cure of chronic HBV infection remains elusive and is rarely achieved with currently available antiviral agents[11]. This situation may be partly due to the presence of drug-resistant mutations of the virus and other factors such as intrinsic stability of the nuclear form of viral genome, the covalently closed circular DNA, and dysfunctional anti-HBV immune response of the host[11,15]. Our findings indicate that drug-resistant mutations of the virus are a minor but important reason for failure of virus eradication, a finding that is consistent with a previous report in which approximately 2% of nucleoside/nucleotide analogue-naïve Chinese patients with chronic hepatitis B had drug-resistant HBV[16].
We are aware of reports that drug resistance can lead to HCC in chronic hepatitis B patients, and high HBV DNA load also can increase the risk of HCC[17]. If HBV DNA becomes negative or decreased after treatment, the risk of HCC decreases, and the risk of HCC is lower with lower HBV DNA loads[18]. Thus, failure to convincingly eradiate HBV in all our patients is a concern, but the magnitude of the risk of HCC developing is unknown.
Based on our results, when the HBV DNA load is lower in the early stages of anti-viral treatment, later outcome is better, and the risk of drug resistance is lower. Others have reported that if patients are treated with standard anti-viral medications according to guidelines and treatment is stopped after successful viral response, 44% had virological recurrence[19] and 50% had clinical recurrence[20]. The reason for recurrence is not known, but an important possibility is that, at the end of treatment, HBV DNA is not suppressed to an adequate level. We believe that the lower the HBV DNA load, the better the prognosis. High-sensitivity HBV DNA detection is useful in predicting anti-viral efficacy as well as in monitoring viral replication and recurrence after cessation of treatment. Patients whose HBV DNA is ≥ 10 IU/mL should be closely monitored, and drug-resistant loci tested when necessary, so that the treatment regimen can be adjusted at an appropriate time.
Our study has some limitations. First, the patient population was from a specific region of China; further studies are needed to determine whether the present results are applicable to broader populations. Second, there may have been a patient-selection bias, as financial consideration may have affected the choice of therapy. Third, patients’ compliance to the prescribed medications was not assessed; thus, it is possible that some patients did not respond to therapy because of noncompliance. Despite the study’s limitations, it expands our knowledge of the therapeutic response to chronic HBV infection in a field still filled with uncertainties about the most efficacious drug regimens and duration of treatment.
The HBV DNA load at 24 wk of antiviral treatment appeared to be a valid predictor of the response rate at 96 wk in patients with chronic hepatitis B. Patients with a lower DNA load at 24 wk had a low DNA load and a higher response rate at 96 wk. Similarly, HBeAg negative conversion and seroconversion rates were more favorable in patients with very low HBV DNA loads at 24 wk than in those with higher DNA loads. ROC curve analysis revealed that the predictability of two-year antiviral treatment efficacy was better in patients with a low initial DNA load than in patients with a higher DNA load at 24 wk. Finally, DNA sequence analysis revealed that some patients who failed to respond to anti-HBV therapy probably had drug-resistant mutations. Results of this study can help in optimizing antiviral therapy in chronic hepatitis B.
Chronic hepatitis B is a human viral liver disease that is especially challenging because of its resistance to treatment and proclivity for progression to chronic liver disease with serious manifestations. Although effective and safe immunization to prevent hepatitis B viral infection is available, hepatitis B virus (HBV) continues to pose a major threat to human health worldwide. An estimated 350 million people have had chronic HBV infection. Universal agreement for the treatment of chronic hepatitis B has not been achieved. Newer medications, i.e., orally administered nucleoside (acid) analogues, strongly inhibit HBV and have been extensively used in clinical practice. However, the optimal course of nucleoside analogue therapy remains uncertain. Moreover, drug resistance may develop after long-term administration of nucleoside analogues.
Complete eradication of the virus is a goal in treatment of HBV infection. However, this goal is rarely achieved with available antiviral agents. Therapeutic regimens to reach optimal outcomes are being explored.
In our research, the authors aimed to determine if the amount of hepatitis B viral DNA present in patients’ blood after 24 wk of treatment with nucleoside analogues would accurately predict the amount of virus present at 96 wk. This information would help determine the patients’ need for continued treatment and their possible long-term outcomes. The authors evaluated 172 Chinese patients who were newly diagnosed with chronic hepatitis B and were treated with nucleoside analogues. They found that indeed the hepatitis B viral DNA load at 24 wk of antiviral treatment was a valid predictor of the response rate at 96 wk in patients with chronic hepatitis B. The findings are consistent with but more extensive than results of few published studies on the time course of response to treatment of chronic hepatitis B virus infection treated with nucleoside (acidic) analogues.
The new information derived from our study will help in optimizing antiviral therapy in chronic hepatitis B. It will help in determining if medical treatment for 24 wk is adequate or whether treatment for longer than 96 wk will be needed.
Most readers will understand the terms used in this study. They may wish to know that nucleoside analogues are an important class of antiviral agents now commonly used in the therapy of human immunodeficiency virus infection, HBV, cytomegalovirus, and herpes simplex virus infection.
A very good article, suitable for publication. The research methodology is nice, the article is well written and clear, and the conclusions accord with the results.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Jeng WJ, Rakhshan V S- Editor: Qi Y L- Editor: Wang TQ E- Editor: Wang CH
| 1. | Pan CQ, Zhang JX. Natural History and Clinical Consequences of Hepatitis B Virus Infection. Int J Med Sci. 2005;2:36-40. [PubMed] [Cited in This Article: ] |
| 2. | Dienstag JL. Hepatitis B virus infection. N Engl J Med. 2008;359:1486-1500. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 689] [Cited by in F6Publishing: 663] [Article Influence: 41.4] [Reference Citation Analysis (0)] |
| 3. | Ganem D, Prince AM. Hepatitis B virus infection--natural history and clinical consequences. N Engl J Med. 2004;350:1118-1129. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1700] [Cited by in F6Publishing: 1653] [Article Influence: 82.7] [Reference Citation Analysis (0)] |
| 4. | Iloeje UH, Yang HI, Su J, Jen CL, You SL, Chen CJ. Predicting cirrhosis risk based on the level of circulating hepatitis B viral load. Gastroenterology. 2006;130:678-686. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1164] [Cited by in F6Publishing: 1101] [Article Influence: 61.2] [Reference Citation Analysis (0)] |
| 5. | European Association For The Study Of The Liver. EASL clinical practice guidelines: Management of chronic hepatitis B virus infection. J Hepatol. 2012;57:167-185. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2323] [Cited by in F6Publishing: 2339] [Article Influence: 194.9] [Reference Citation Analysis (0)] |
| 6. | Liaw YF, Kao JH, Piratvisuth T, Chan HL, Chien RN, Liu CJ, Gane E, Locarnini S, Lim SG, Han KH. Asian-Pacific consensus statement on the management of chronic hepatitis B: a 2012 update. Hepatol Int. 2012;6:531-561. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 742] [Cited by in F6Publishing: 759] [Article Influence: 63.3] [Reference Citation Analysis (0)] |
| 7. | Liaw YF, Gane E, Leung N, Zeuzem S, Wang Y, Lai CL, Heathcote EJ, Manns M, Bzowej N, Niu J. 2-Year GLOBE trial results: telbivudine Is superior to lamivudine in patients with chronic hepatitis B. Gastroenterology. 2009;136:486-495. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 435] [Cited by in F6Publishing: 468] [Article Influence: 31.2] [Reference Citation Analysis (0)] |
| 8. | Zeuzem S, Gane E, Liaw YF, Lim SG, DiBisceglie A, Buti M, Chutaputti A, Rasenack J, Hou J, O’Brien C. Baseline characteristics and early on-treatment response predict the outcomes of 2 years of telbivudine treatment of chronic hepatitis B. J Hepatol. 2009;51:11-20. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 161] [Cited by in F6Publishing: 172] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 9. | Zoulim F, Białkowska-Warzecha J, Diculescu MM, Goldis AE, Heyne R, Mach T, Marcellin P, Petersen J, Simon K, Bendahmane S. Entecavir plus tenofovir combination therapy for chronic hepatitis B in patients with previous nucleos(t)ide treatment failure. Hepatol Int. 2016;10:779-788. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 13] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 10. | Liang X, Fan R, Sun J, Shaikh J, Taneja A, Gupta S, Hamed K. Effect of Telbivudine Versus Other Nucleos(t)ide Analogs on HBeAg Seroconversion and Other Outcomes in Patients with Chronic Hepatitis B: A Network Meta-Analysis. Adv Ther. 2016;33:519-531. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 8] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 11. | Chang J, Guo F, Zhao X, Guo JT. Therapeutic strategies for a functional cure of chronic hepatitis B virus infection. Acta Pharm Sin B. 2014;4:248-257. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 42] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 12. | Zhu H, Wang C, Zhang Y, Wei S, Li X, Zhang Z. Prediction model for sustained hepatitis B e antigen seroconversion to peginterferon alfa-2a in chronic hepatitis B. J Gastroenterol Hepatol. 2016;31:1963-1970. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 11] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 13. | Martinot-Peignoux M, Lapalus M, Maylin S, Boyer N, Castelnau C, Giuily N, Pouteau M, Moucari R, Asselah T, Marcellin P. Baseline HBsAg and HBcrAg titres allow peginterferon-based ‘precision medicine’ in HBeAg-negative chronic hepatitis B patients. J Viral Hepat. 2016;23:905-911. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 38] [Cited by in F6Publishing: 29] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 14. | van Bömmel F, Bartens A, Mysickova A, Hofmann J, Krüger DH, Berg T, Edelmann A. Serum hepatitis B virus RNA levels as an early predictor of hepatitis B envelope antigen seroconversion during treatment with polymerase inhibitors. Hepatology. 2015;61:66-76. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 174] [Cited by in F6Publishing: 175] [Article Influence: 19.4] [Reference Citation Analysis (0)] |
| 15. | Lin CL, Kao JH. Review article: novel therapies for hepatitis B virus cure - advances and perspectives. Aliment Pharmacol Ther. 2016;44:213-222. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 39] [Cited by in F6Publishing: 35] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 16. | Li X, Liu Y, Zhao P, Wang Y, Chen L, Xin S, Zhang XX, Xu D. Investigation into drug-resistant mutations of HBV from 845 nucleoside/nucleotide analogue-naive Chinese patients with chronic HBV infection. Antivir Ther. 2015;20:141-147. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 14] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 17. | Chan HL, Tse CH, Mo F, Koh J, Wong VW, Wong GL, Lam Chan S, Yeo W, Sung JJ, Mok TS. High viral load and hepatitis B virus subgenotype ce are associated with increased risk of hepatocellular carcinoma. J Clin Oncol. 2008;26:177-182. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 226] [Cited by in F6Publishing: 225] [Article Influence: 14.1] [Reference Citation Analysis (1)] |
| 18. | Ikeda K, Arase Y, Kobayashi M, Someya T, Hosaka T, Saitoh S, Sezaki H, Akuta N, Suzuki F, Suzuki Y. Hepatitis B virus-related hepatocellular carcinogenesis and its prevention. Intervirology. 2005;48:29-38. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 20] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 19. | Fung J, Lai CL, Chan SC, But D, Seto WK, Cheng C, Wong DK, Lo CM, Fan ST, Yuen MF. Correlation of liver stiffness and histological features in healthy persons and in patients with occult hepatitis B, chronic active hepatitis B, or hepatitis B cirrhosis. Am J Gastroenterol. 2010;105:1116-1122. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 44] [Cited by in F6Publishing: 46] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 20. | Liang Y, Jiang J, Su M, Liu Z, Guo W, Huang X, Xie R, Ge S, Hu J, Jiang Z. Predictors of relapse in chronic hepatitis B after discontinuation of anti-viral therapy. Aliment Pharmacol Ther. 2011;34:344-352. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 97] [Cited by in F6Publishing: 73] [Article Influence: 5.6] [Reference Citation Analysis (0)] |