Published online Mar 21, 2017. doi: 10.3748/wjg.v23.i11.1944
Peer-review started: December 29, 2016
First decision: January 19, 2017
Revised: February 7, 2017
Accepted: March 2, 2017
Article in press: March 2, 2017
Published online: March 21, 2017
Inflammatory bowel disease (IBD) results from a complex series of interactions between susceptibility genes, the environment, and the immune system. Recently, some studies provided strong evidence that the process of autophagy affects several aspects of mucosal immune responses. Autophagy is a cellular stress response that plays key roles in physiological processes, such as innate and adaptive immunity, adaptation to starvation, degradation of aberrant proteins or organelles, antimicrobial defense, and protein secretion. Dysfunctional autophagy is recognized as a contributing factor in many chronic inflammatory diseases, including IBD. Autophagy plays multiple roles in IBD pathogenesis by altering processes that include intracellular bacterial killing, antimicrobial peptide secretion by Paneth cells, goblet cell function, proinflammatory cytokine production by macrophages, antigen presentation by dendritic cells, and the endoplasmic reticulum stress response in enterocytes. Recent studies have identified susceptibility genes involved in autophagy, such as NOD2, ATG16L1, and IRGM, and active research is ongoing all over the world. The aim of this review is a systematic appraisal of the current literature to provide a better understanding of the role of autophagy in the pathogenesis of IBD. Understanding these mechanisms will bring about new strategies for the treatment and prevention of IBD.
Core tip: Recent studies provide strong evidence that the process of autophagy affects several aspects of mucosal immune responses. Autophagy is a cellular stress response that plays key roles in physiological processes. Dysfunctional autophagy is recognized as a contributing factor in many chronic inflammatory diseases, including inflammatory bowel disease (IBD). Autophagy plays multiple roles in IBD pathogenesis. Recent studies have identified susceptibility genes involved in autophagy, such as NOD2, ATG16L1, and IRGM, and active research is ongoing around the world. The aim of this review is a systematic appraisal of current literature to provide a better understanding of the role of autophagy in IBD pathogenesis.
- Citation: Iida T, Onodera K, Nakase H. Role of autophagy in the pathogenesis of inflammatory bowel disease. World J Gastroenterol 2017; 23(11): 1944-1953
- URL: https://www.wjgnet.com/1007-9327/full/v23/i11/1944.htm
- DOI: https://dx.doi.org/10.3748/wjg.v23.i11.1944
Inflammatory bowel disease (IBD) is a chronic inflammatory disease involving idiopathic inflammation, mainly in the gastrointestinal tract; defined more specifically, it comprises ulcerative colitis (UC) and Crohn’s disease (CD). Both are characterized by onset at a young age, and the number of affected patients has risen sharply in recent years in Europe and the United States, as well as in Japan[1]. Thus, there is a pressing need to understand their pathologies and create effective treatments. Researchers, mainly in Europe and the United States, have been trying to identify disease-susceptibility genes for IBD. Nucleotide-binding oligomerization domain-containing protein 2 (NOD2) was the first susceptibility gene identified for CD[2,3], and in recent years genome-wide association studies (GWAS) have made it possible to perform comprehensive searches for susceptibility genes. In 2007, autophagy-related 16-like 1 (ATG16L1) was identified as an autophagy-related gene[4]. This was the first study to show a relationship between autophagy and a specific disease. Since then, the role of autophagy in the pathogenesis of IBD has been investigated all over the world.
This review will evaluate the current literature to provide a better understanding of the role of autophagy in the pathophysiology of IBD.
The gastrointestinal tract not only absorbs fluid and nutrients, but is constantly involved in regulating and maintaining the gut flora, immune responses to food antigens and other substances, and homeostasis. IBD occurs when this homeostasis is impaired. Recent research has shown that IBD is caused by chronic intestinal inflammation, which occurs because of gene variations that can lead to disease susceptibility, changes in the structure of the intestinal flora needed to maintain intestinal homeostasis, and abnormal intestinal mucosal immune responses[5-7].
The role of genetic factors in IBD has been previously reported[8], and several researchers are seeking disease-susceptibility genes and trying to find customized treatments for individual patients[9]. To date, approximately 200 loci have been identified as being associated with both forms of IBD. Within these 200 loci, based upon single nucleotide polymorphism frequencies in IBD subjects versus controls, are approximately 1500 potential associated genes[10,11]. Representative autophagy-related genes are NOD2, ATG16L1, and immunity related guanosine triphosphatase M (IRGM)[1-3,12]. Autophagy has been linked to a variety of diseases, but its link to IBD is currently the subject of much debate.
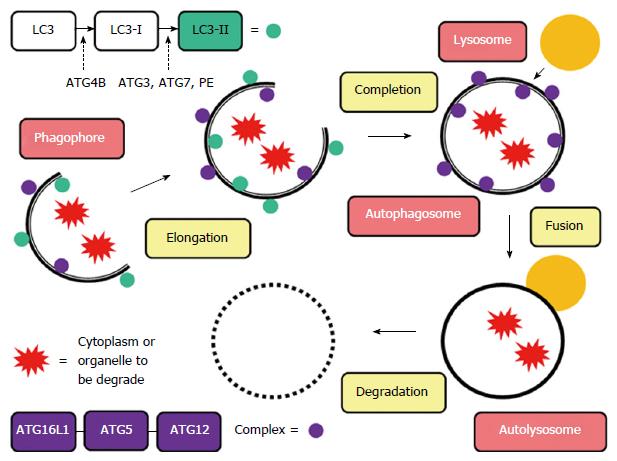
Autophagy (from the Greek “auto” oneself and “phagy” to eat) refers to any cellular degradative pathway that involves the delivery of cytoplasmic cargo to the lysosome. During this process, the endoplasmic reticulum or other membranous cellular structures respond to stimuli by generating a double-membrane structure called a phagophore. On this phagophore, ATG16L1 forms a complex with an ATG5-ATG12 conjugate, which multimerizes and then lipidates LC3 (LC3-II). Simultaneously, the phagophore elongates to envelop the cytoplasm or organelle to be degraded, forming an autophagosome, a unique double-membrane organelle. The outer membrane of the autophagosome then fuses with a lysosome to form an autolysosome, and the inner membrane degrades and absorbs its contents (Figure 1)[13-15]. This process, along with the ubiquitin proteasome pathway (UPP) system, triggers the intracellular protein degradation mechanism. The process is also responsible for mechanisms such as adaptation to starvation, defense against infections, carcinogenesis, antigen presentation, and quality control of intracellular proteins. It maintains appropriate cellular homeostasis and provides the structural processes necessary for organ renewal[16]. Yet, unlike the UPP system, autophagy is also able to degrade mitochondria and other organelles.
A remarkable analysis of autophagy-related factor groups showed that, in addition to its role in metabolism, autophagy plays an important role in the innate immune response[13]. Innate immunity is a mechanism by which nearly all multicellular organisms protect themselves from pathogens. Innate immunity signaling pathways are activated when the structural patterns of a pathogen’s components are recognized (i.e., the cell wall components of a bacterium or the genome of a virus). As noted above, autophagy was initially considered to be a nonspecific mechanism for degrading substances by incorporating them into a membrane structure, but recent research has shown that autophagosomes selectively isolate a variety of substrates[17]. However, besides autophagy of pathogens (xenophagy)[18,19] and autophagy of damaged mitochondria (mitophagy)[20,21], very little is understood about which substrates autophagy degrades when it functions as part of innate immunity.
Autophagic dysfunction causes several diseases[22-24], among which CD is being most extensively researched. The above mentioned GWAS found several genetic variants linked to CD onset, such as NOD2 and ATG16L1. A summary of these variants is given below (Table 1).
| Gene | Chromosomal site | Relation to autophagy |
| NOD2 | 16q12.1 | Intracellular bacterial sensing |
| Autophagosome formation | ||
| ATG16L1 | 2p37.1 | Autophagosome formation |
| Suppressing Paneth cells | ||
| IRGM | 5q33.1 | Phagosome maturation |
| Virus-induced autophagy | ||
| IL-23R | 1p31.3 | Through effects on IL-1 secretion |
| XIAP | Xq25 | Physiological inhibitor of autophagy |
| LRRK2 | 12q12 | Autophagosomal-lysosomal degradation |
| ULK1 | 12q24.33 | Regulated by TORC1 and AMPK |
| VDR | 12q13.11 | Regulate the expression of NOD2 |
| MTMR3 | 22q12.2 | Autophagosome formation |
NOD2, located on chromosome 16q12.1, was the first disease-susceptibility gene discovered for CD. Its genetic variants are common in European and American patients, but have not been found in Asian patients. NOD2 is a pattern-recognition receptor that is involved in the homeostasis of intestinal immunity. It acts through mechanisms like autophagy, intracellular bacterial sensing, controlling the expression of the antibacterial peptide α-defensin in the Paneth cells of the small intestine, and improving immune tolerance by suppressing toll-like receptor (TLR) signals[25]. NOD2 recruits the autophagy protein ATG16L1 to the plasma membrane at the bacterial entry site; mutant NOD2 failed to recruit ATG16L1 to the plasma membrane and wrapping of invading bacteria by autophagosomes was impaired. Therefore, patients with CD with NOD2 variants are considered to exhibit disorders of autophagy[26-28]. When the mechanism of autophagy is impaired, lipopolysaccharides and damage-associated molecular patterns trigger signaling by stimulating TLR and NOD-like receptors, tumor necrosis factor (TNF), and other inflammatory cytokines. They also stimulate caspase-1 causing interleukin (IL)-1β and IL-18 cleavage from precursors, which promotes extracellular secretion (inflammasomes). In an experiment using mice knocked out for ATG16L, which encodes ATG16L1, the protein necessary for the autophagic recruitment, TLR and TNF stimulation led to abnormal inflammasome activity in macrophages and other innate immunity cells[29].
ATG16L1 is a homolog of ATG16 that was first reported by Mizushima et al[30,31]. Along with AT5 and ATG12, this molecule is required to form autophagosomes. Prescott et al[32] reported that the incidence of CD was likely to be two times higher in people with the T300A variant, an ATG16L1 variant with a threonine-to-alanine substitution at amino-acid position 300. Later, a meta-analysis of 25 studies showed that T300A caused disease susceptibility to CD[33]. However, no significant difference was observed in an analysis of patients from Japan, South Korea, and China from 25 studies. This suggests that European and American patients exhibit different genetic factors compared to Asian patients, as is seen with NOD2. Moreover, a meta-analysis of 14 studies on UC reported an odds ratio of 1.06, or almost no difference[33].
The report that ATG16L1 is a CD-susceptibility gene was a groundbreaking discovery suggesting a role for autophagy in the onset of IBD. Since then, several researchers have published studies on the link between ATG16L1 and IBD.
Paneth cells are a specialized type of epithelial cell that are involved in innate immunity in the small intestine. When they come into contact with bacteria or other antigens, these cells release secretory granules containing antimicrobial peptides and a variety of proteins. In 2008, Cadwell et al[34] engineered a mouse with low expression of ATG16L1 (Atg16L1HM mouse). Tissue analysis did not find lysozymes that are normally seen in the ileal mucosa, but found abnormal Paneth cell granule secretion. Moreover, they analyzed Paneth cells in non-inflamed areas of the ileum in patients with CD homozygous for the ATG16L1 variant T300A, and found abnormal Paneth cells that strongly resembled those observed in Atg16L1HM mice. This suggests that ATG16L1 may also play an important role by suppressing Paneth cells in humans. In a relatively recent study, Lassen et al[35] generated a knock-in mouse model expressing ATG16L1T300A. Such mice do not develop spontaneous inflammation, although they exhibit morphological defects in both Paneth cells and goblet cells. Furthermore, the presence of the T300A mutation in ATG16L1 leads to aberrant functionality of Paneth cells. These findings indicate the reason there is believed to be a close relationship between ATG16L1 variants and Paneth cells.
Further, Murthy et al[36] reported that ATG16L1 amino-acid positions 296 to 299 form a caspase cleavage motif, which greatly increases ATG16L1 sensitivity when the cellular stress response activates caspase-3 in the presence of the T300A variant. This may result in impaired autophagy, leading to CD onset, and suggests that ATG16L1 plays a role at the molecular level in CD onset.
In 2010, Cadwell et al[37] reported interesting data on role of ATG16L1 by using Atg16L1HM mice infected with MNV CR6, a species of mouse norovirus. MNV CR6-infected Atg16L1HM mice showed abnormal secretion of Paneth cell granules, similar to that described above. This was not observed in wild-type mice without an ATG16L1 variant, or in mice infected with a different MNV strain or with inactivated MNV. Administration of dextran sulfate sodium (DSS) to these infected mice led to pathology similar to that observed in human patients with CD: inflammation extending to the muscle layer and mesentery, and atrophy of the ileal villi, neither of which has been previously reported with DSS colitis. These symptoms were significantly suppressed by administering TNF-α antibodies or antibiotics. A recent report suggested that ATG16L1 polymorphisms promote disease through defects in “sensing” protective signals from the microbiome, defining a potentially critical gene-environment etiology for IBD[38].
These data suggest that in addition to ATG16L1 variants, CD onset is influenced by a complex variety of environmental factors, including viral infections and enterobacteria.
In a 2007 GWAS, Parkes et al[39] reported that the IRGM gene on chromosome 5q33.1 was a CD-susceptibility gene. In humans, IRGM is a 20 kDa protein formed from 181 amino acids that is expressed in the large intestine, small intestine, and lymphocytes. IRGM is related to bacterial killing, vacuolar trafficking and acidification, phagosome maturation, and virus-induced autophagy. Moreover, it is known to be involved in controlling intracellular Mycobacterium tuberculosis by autophagy in macrophages[40]. A small nuclear polymorphism (SNP) with susceptibility is adjacent to IRGM, but detailed sequencing of IRGM did not reveal any CD-related variants with modified amino acids. This suggests the possibility that changes of IRGM expression, transcript splicing, or the ratio of translation of the protein are related to the development of CD.
In 2008, McCarroll et al[41] discovered a 20 kb deletion polymorphism upstream from IRGM that was linked to an SNP correlating with CD. In addition, they reported that the expression of IRGM suppressed autophagy of intracellular bacteria, which has been linked to CD, suggesting a role in the pathology of CD.
Recently, Rufini et al[42] reported that IRGM polymorphisms were important for CD susceptibility and phenotype modulation (fibrostricturing behavior, ileal disease, perianal disease, and intestinal resection).
IL-23 is a heterodimeric cytokine produced by activated macrophages and dendritic cells. It consists of two subunits, a p40 subunit, shared with IL-12, and a specific IL-23 subunit called p19[43,44]. It has been shown that IL-23 is involved in the initiation of the innate and adaptive immune activation that characterizes IBD. It binds a complex of IL-23 receptor (IL-23R) and IL-12Rβ subunits. IL-23R is predominantly expressed on activated/memory T cells, T-cell clones, natural killer cells and, at low levels, in monocytes, macrophages, and dendritic cell populations[45,46]. Recent studies have shown association of the IL-23R gene with chronic inflammatory diseases, especially IBD[47,48]. It is also reported that autophagy regulates IL-23 secretion and innate T cell responses through effects on IL-1 secretion[49].
XIAP (X-linked inhibitor of apoptosis) is one of several inhibitor of apoptosis proteins (IAPs). IAPs were initially identified in baculoviruses, where they prevent defensive apoptosis of host cells[50]. Among the mammalian IAPs, XIAP is the most extensively studied and best characterized. XIAP has the most potent anti-apoptotic ability[51], which is believed to be primarily related to direct binding and inhibiting of caspases, the apoptotic proteases that are responsible for the initiation and execution of apoptosis[52]. Huang et al[53] showed that XIAP is a physiological inhibitor of autophagy, and has been associated with a variety of diseases that have been linked to autophagy. XIAP is related to X-linked lymphoproliferative syndrome type 2 (XLP2), a type of primary immunodeficiency. However, a genetic analysis performed by Zeissig et al[54] found XIAP variants in only 4% of male patients with childhood-onset CD. Recently, Schwerd et al[55] showed impaired antibacterial autophagy links granulomatous intestinal inflammation in Niemann-Pick disease type C1 and XIAP deficiency with NOD2 variants in CD.
LRRK2 (leucine-rich repeat kinase 2) is a large multidomain protein belonging to the ROCO family of proteins, which are characterized by the presence of leucine-rich repeats, a Ras of complex (ROC) GTPase domain, a C-terminal ROC linker region, and a kinase domain. LRRK2 localizes to specific membrane subdomains, including endolysosomal structures in many kinds of cells. Studies showed that LRRK2 KO mice displayed an increase in the number and size of secondary lysosomes and autolysosome-like structures. Abnormal accumulation of undigested material indicates an impairment in the autophagosomal-lysosomal degradation system (autophagy-lysosomal clearance pathway).
LRRK2 has been identified as a disease-susceptibility gene for Parkinson’s disease, leprosy, and CD. The CD-associated SNP is located upstream of the coding sequence of LRRK2[56,57]. It was reported that LRRK2 expression levels were found to be significantly upregulated in colonic biopsy specimens from inflamed tissues of patients with CD[58]. LRRK2 is known to be expressed only in mucosal lymphocytes in the colonic mucosa, but little else is known about it.
ULK1 (Unc-51 like autophagy activating kinase 1) is one of the key regulators of autophagy initiation and progression. Mammals have two homologs of the yeast autophagy-initiating ATG1 kinase, ULK1 and ULK2. ULK1 is regulated by the nutrient- and energy-sensitive kinases TORC1 and AMPK. The tight regulation of ULK activity by intracellular energy and nutrient levels is in keeping with a central role for autophagy in the protection of cells from starvation.
Henckaerts et al[59] selected human homologs of 12 yeast autophagy genes, known to be found in IBD-related loci from GWAS, and conducted a meta-analysis of these searches. An analysis of correlations with CD identified ULK1 as a CD-susceptibility gene. ULK1 activity is regulated by a complex array of multiple phosphorylation and dephosphorylation events that influence the binding of regulatory and effector autophagy proteins[60,61]. However, little is known about the action of ULK1 in association with IBD, and further research is necessary.
VDR (the vitamin D receptor) regulates the expression of NOD2, and it has been suggested that it controls the mechanism of autophagy. Unlike other genes, VDR has been shown to be a UC-susceptibility gene, not only among Europeans and Americans, but also in Asian and Middle Eastern populations[62]. Vitamin D deficiency increases the risk of CD onset[63]; thus, analyzing its signaling pathways could help elucidate the pathology of this disease.
Recently, Wu et al[64] showed a fundamental relationship between the VDR, autophagy, and gut microbial assemblage that is essential for maintaining intestinal homeostasis, but also contributes to the pathophysiology of IBD. Furthermore, Abreu-Delgado et al[65] reported that levels of serum vitamin D correlate positively with colonic VDR expression in visually normal mucosa; whereas inflammation correlates negatively with colonic VDR expression in visually diseased mucosa. The VDR needs further research.
MTMR3 (myotubularin-related protein 3) plays a role in autophagosome formation[66]. The myotubularin family is a class of PI3-phosphatases that regulate several physiological and pathophysiological phenomena, including endosomal trafficking, apoptosis, autophagy, and muscle development. As a member of this family, MTMR3 has been considered to play a negative role in the initiation stage of autophagy. Recent reports indicate that MTMR3 has at least two opposite functions in the autophagy pathway, inhibition of mechanistic target of rapamycin complex 1 (mTORC1) and reduction of local PI3P levels[67,68]. In this regard, the function of MTMR3 in autophagy remains unclear.
Widely used therapeutic agents for IBD include steroids and 5-aminosalicylic acid (5-ASA), as well as immunoregulatory drugs such as azathioprine, and biologicals such as anti-TNF-α formulations. The process of autophagy is closely related to each of these existing therapeutic agents. The following sections summarize these relationships (Table 2).
| Drug | Influence on autophagy | Mechanism related to autophagy |
| 5-ASA | Promotion | Through NF-κB signaling pathway |
| Corticosteroid | Promotion | Through NF-κB signaling pathway |
| Through mTORC1 signaling pathway | ||
| Through overexpression of Bcl-2 | ||
| in immature T-lymphocytes | ||
| Osteocyte viability | ||
| Thiopurine | Promotion | Clearance of TPMT*3A aggregates |
| (AZA, 6-MP) | and/or aggregate precursors | |
| Protective role in hepatocytes | ||
| Immunomodulatory drugs | Promotion | Response to toxicity |
| (CsA, FK506) | Through mTORC1 signaling pathway | |
| Biological drugs | Inhibition | Anti-TNF agents inhibit |
| (IFX, ADA, etc.) | autophagy (not yet clear) |
The mechanism of action of 5-ASA has been described in several studies. The suppression of peroxisome proliferator-activated receptor gamma (PPARγ) due to the production of inflammatory cytokines is said to contribute to the intestinal inflammation seen in patients with IBD[69]. 5-ASA is considered to exert its anti-inflammatory action by acting on PPARγ in epithelial cells, and by regulating signal transmission from NF-κB and TLR[70]. Considering that NF-κB signaling is associated with autophagy[71], it might be that 5-ASA indirectly regulates autophagy.
The first-line treatment to induce remission for CD and UC is often corticosteroids. Corticosteroids downregulate proinflammatory cytokines, including IL-1, IL-6, and TNFα. Furthermore, inflammatory signaling induced by NF-κB is decreased by interaction with corticosteroid receptors[72], and, as noted above, NF-κB signaling regulates autophagy[71]. It has also been shown that corticosterone treatment affects mTORC1 signaling pathways[73]. It was reported that mTORC1 pathways and autophagy play an important role in the response to treatment with corticosteroids[74]. Corticosteroids are able to induce apoptosis in immature T lymphocytes, as these cells lack the inhibitor of apoptosis protein Bcl-2. It has been shown that overexpression of Bcl-2 in immature T lymphocytes can increase autophagy levels, presumably due to inhibition of apoptosis[75].
A relationship between corticosteroids and autophagy has been observed, not only for their therapeutic effects, but also for the adverse effects that accompany treatment. It has been shown, both in vitro and in vivo, that low doses of prednisolone and dexamethasone induce autophagy in osteocytes, and this is associated with osteocyte viability[76,77]. However, higher doses of corticosteroids induce apoptosis, suggesting that autophagy may act as a protective mechanism against the cytotoxic effects of corticosteroids[76].
Thiopurines, including azathioprine and 6-mercaptopurine, are immunosuppressant drugs used to maintain remission in patients with IBD[78]. Thiopurines and autophagy have also been shown to be correlated by the adverse effects of treatment. The thiopurine S-methyltransferase (TPMT) genetic polymorphism is important for thiopurine metabolism. Individuals with inherited decreases in TPMT activity, mainly as a result of the effects of the TPMT*3A allele (minor allele frequency in Caucasians of approximately 5%)[79], are at greatly increased risk for severe life-threatening myelosuppression when treated with “standard” doses of thiopurine drugs[80-83]. It was shown that autophagy might represent an important route for the clearance of TPMT*3A aggregates and/or aggregate precursors[84]. Due to the severe adverse effects of thiopurines, a potential protective role for autophagy in hepatocytes has been investigated; it has been shown that autophagy has a protective role in hepatocytes during thiopurine therapy[78].
Cyclosporine A (CsA), FK506, and methotrexate (MTX) are immunomodulatory drugs used mainly as second-line treatments to induce and maintain remission in severe, steroid-refractory CD[85], with more recent evidence suggesting a role for FK506 in UC[86]. Although some evidence suggests that CsA and FK506 are involved in autophagy, no relationship has been identified between MTX and autophagy.
Several studies have shown that treatment with CsA can induce autophagy in response to toxicity (such as CsA-induced nephrotoxicity), either as a survival process or as part of a cell death mechanism[87-89].
FK506 inhibits calcineurin by forming a complex with the immunophilin FK506 binding protein 12 (FKBP12), which is involved in immunoregulation[90]. FKBP12 is also the direct target of rapamycin, an inhibitor of mTORC1. The molecular mechanism by which mTORC1 regulates autophagy in mammals is being investigated[91,92], while future research is expected to help understand the relationship between FK506 and autophagy.
The most commonly used biological drug for IBD is the anti-TNFα antibody infliximab. Other anti-TNFα treatments approved for treatment of patients with IBD patients include adalimumab, golimumab for UC only, and certolizumab pegol. Anti-TNFα biosimilars have also recently been developed[93]. The relationship between TNFα and autophagy has been confirmed in synovial fibroblasts[94], skeletal muscles[95], and trophoblastic cells[96]. These studies suggest that anti-TNF agents would inhibit autophagy, and while the mechanism of action is not yet completely clear, it has been the subject of extensive research lately.
The above data summarizes the relationship between autophagy and various drugs. However, existing medical therapies do not relieve the symptoms in many patients, and surgical intervention is often necessary. There is, therefore, a pressing need to develop new therapeutic agents. As seen in this review, autophagy plays an important role in controlling the immune system; hence drugs that regulate autophagy have received much attention as potential new therapeutic targets for IBD[97]. Further investigation of the role of autophagy in existing IBD therapies, and development of new therapeutic agents regulating autophagy, are the needs of the hour.
GWAS has identified several disease-susceptibility genes, and studies on the pathology and etiology of IBD are being regularly published; however, more aspects of IBD pathogenesis should be clarified. As the number of patients with IBD is still increasing around the world, particularly among the young, it is essential that the mechanism of IBD is elucidated and treatments based on this mechanism are developed. A better understanding of the relationship between autophagy and IBD will result in better IBD therapy in future.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: Japan
Peer-review report classification
Grade A (Excellent): A
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Bezmin Abadi AT, de Almeida Araujo EJ, Patial V S- Editor: Qi Y L- Editor: A E- Editor: Zhang FF
| 1. | Molodecky NA, Soon IS, Rabi DM, Ghali WA, Ferris M, Chernoff G, Benchimol EI, Panaccione R, Ghosh S, Barkema HW. Increasing incidence and prevalence of the inflammatory bowel diseases with time, based on systematic review. Gastroenterology. 2012;142:46-54.e42; quiz e30. [PubMed] [DOI] [Cited in This Article: ] |
| 2. | Hugot JP, Chamaillard M, Zouali H, Lesage S, Cézard JP, Belaiche J, Almer S, Tysk C, O’Morain CA, Gassull M. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature. 2001;411:599-603. [PubMed] [DOI] [Cited in This Article: ] |
| 3. | Ogura Y, Bonen DK, Inohara N, Nicolae DL, Chen FF, Ramos R, Britton H, Moran T, Karaliuskas R, Duerr RH. A frameshift mutation in NOD2 associated with susceptibility to Crohn’s disease. Nature. 2001;411:603-606. [PubMed] [DOI] [Cited in This Article: ] |
| 4. | Hampe J, Franke A, Rosenstiel P, Till A, Teuber M, Huse K, Albrecht M, Mayr G, De La Vega FM, Briggs J. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat Genet. 2007;39:207-211. [PubMed] [DOI] [Cited in This Article: ] |
| 5. | Kaser A, Blumberg RS. Autophagy, microbial sensing, endoplasmic reticulum stress, and epithelial function in inflammatory bowel disease. Gastroenterology. 2011;140:1738-1747. [PubMed] [DOI] [Cited in This Article: ] |
| 6. | Kaser A, Zeissig S, Blumberg RS. Inflammatory bowel disease. Annu Rev Immunol. 2010;28:573-621. [PubMed] [DOI] [Cited in This Article: ] |
| 7. | Khor B, Gardet A, Xavier RJ. Genetics and pathogenesis of inflammatory bowel disease. Nature. 2011;474:307-317. [PubMed] [DOI] [Cited in This Article: ] |
| 8. | Weterman IT, Peña AS. Familial incidence of Crohn’s disease in The Netherlands and a review of the literature. Gastroenterology. 1984;86:449-452. [PubMed] [Cited in This Article: ] |
| 9. | Fiocchi C. Tailoring Treatment to the Individual Patient - Will Inflammatory Bowel Disease Medicine Be Personalized? Dig Dis. 2015;33 Suppl 1:82-89. [PubMed] [DOI] [Cited in This Article: ] |
| 10. | Cho JH, Brant SR. Recent insights into the genetics of inflammatory bowel disease. Gastroenterology. 2011;140:1704-1712. [PubMed] [DOI] [Cited in This Article: ] |
| 11. | Ek WE, D’Amato M, Halfvarson J. The history of genetics in inflammatory bowel disease. Ann Gastroenterol. 2014;27:294-303. [PubMed] [Cited in This Article: ] |
| 12. | Goldstein DB. Common genetic variation and human traits. N Engl J Med. 2009;360:1696-1698. [PubMed] [DOI] [Cited in This Article: ] |
| 13. | Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, Lee JC, Schumm LP, Sharma Y, Anderson CA, Essers J, Mitrovic M, Ning K, Cleynen I, Theatre E, Spain SL, Raychaudhuri S, Goyette P, Wei Z, Abraham C, Achkar JP, Ahmad T, Amininejad L, Ananthakrishnan AN, Andersen V, Andrews JM, Baidoo L, Balschun T, Bampton PA, Bitton A, Boucher G, Brand S, Büning C, Cohain A, Cichon S, D’Amato M, De Jong D, Devaney KL, Dubinsky M, Edwards C, Ellinghaus D, Ferguson LR, Franchimont D, Fransen K, Gearry R, Georges M, Gieger C, Glas J, Haritunians T, Hart A, Hawkey C, Hedl M, Hu X, Karlsen TH, Kupcinskas L, Kugathasan S, Latiano A, Laukens D, Lawrance IC, Lees CW, Louis E, Mahy G, Mansfield J, Morgan AR, Mowat C, Newman W, Palmieri O, Ponsioen CY, Potocnik U, Prescott NJ, Regueiro M, Rotter JI, Russell RK, Sanderson JD, Sans M, Satsangi J, Schreiber S, Simms LA, Sventoraityte J, Targan SR, Taylor KD, Tremelling M, Verspaget HW, De Vos M, Wijmenga C, Wilson DC, Winkelmann J, Xavier RJ, Zeissig S, Zhang B, Zhang CK, Zhao H; International IBD Genetics Consortium (IIBDGC), Silverberg MS, Annese V, Hakonarson H, Brant SR, Radford-Smith G, Mathew CG, Rioux JD, Schadt EE, Daly MJ, Franke A, Parkes M, Vermeire S, Barrett JC, Cho JH. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119-124. [PubMed] [DOI] [Cited in This Article: ] |
| 14. | Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. 2011;469:323-335. [PubMed] [DOI] [Cited in This Article: ] |
| 15. | Hooper KM, Barlow PG, Stevens C, Henderson P. Inflammatory Bowel Disease Drugs: A Focus on Autophagy. J Crohns Colitis. 2017;11:118-127. [PubMed] [DOI] [Cited in This Article: ] |
| 16. | Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27-42. [PubMed] [DOI] [Cited in This Article: ] |
| 17. | Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol. 2011;27:107-132. [PubMed] [DOI] [Cited in This Article: ] |
| 18. | Levine B. Eating oneself and uninvited guests: autophagy-related pathways in cellular defense. Cell. 2005;120:159-162. [PubMed] [DOI] [Cited in This Article: ] |
| 19. | Nozawa T, Minowa-Nozawa A, Aikawa C, Nakagawa I. The STX6-VTI1B-VAMP3 complex facilitates xenophagy by regulating the fusion between recycling endosomes and autophagosomes. Autophagy. 2017;13:57-69. [PubMed] [DOI] [Cited in This Article: ] |
| 20. | Hailey DW, Rambold AS, Satpute-Krishnan P, Mitra K, Sougrat R, Kim PK, Lippincott-Schwartz J. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell. 2010;141:656-667. [PubMed] [DOI] [Cited in This Article: ] |
| 21. | Yao Z, Klionsky DJ. An unconventional pathway for mitochondrial protein degradation. Autophagy. 2016;12:1971-1972. [PubMed] [DOI] [Cited in This Article: ] |
| 22. | Guo R, Lin B, Pan JF, Liong EC, Xu AM, Youdim M, Fung ML, So KF, Tipoe GL. Inhibition of caspase-9 aggravates acute liver injury through suppression of cytoprotective autophagy. Sci Rep. 2016;6:32447. [PubMed] [DOI] [Cited in This Article: ] |
| 23. | Kang YH, Cho MH, Kim JY, Kwon MS, Peak JJ, Kang SW, Yoon SY, Song Y. Impaired macrophage autophagy induces systemic insulin resistance in obesity. Oncotarget. 2016;7:35577-35591. [PubMed] [DOI] [Cited in This Article: ] |
| 24. | Martinez J, Cunha LD, Park S, Yang M, Lu Q, Orchard R, Li QZ, Yan M, Janke L, Guy C. Noncanonical autophagy inhibits the autoinflammatory, lupus-like response to dying cells. Nature. 2016;533:115-119. [PubMed] [DOI] [Cited in This Article: ] |
| 25. | Yano T, Kurata S. Intracellular recognition of pathogens and autophagy as an innate immune host defence. J Biochem. 2011;150:143-149. [PubMed] [DOI] [Cited in This Article: ] |
| 26. | Cooney R, Baker J, Brain O, Danis B, Pichulik T, Allan P, Ferguson DJ, Campbell BJ, Jewell D, Simmons A. NOD2 stimulation induces autophagy in dendritic cells influencing bacterial handling and antigen presentation. Nat Med. 2010;16:90-97. [PubMed] [DOI] [Cited in This Article: ] |
| 27. | Homer CR, Richmond AL, Rebert NA, Achkar JP, McDonald C. ATG16L1 and NOD2 interact in an autophagy-dependent antibacterial pathway implicated in Crohn’s disease pathogenesis. Gastroenterology. 2010;139:1630-1641, 1641.e1-2. [PubMed] [DOI] [Cited in This Article: ] |
| 28. | Travassos LH, Carneiro LA, Ramjeet M, Hussey S, Kim YG, Magalhães JG, Yuan L, Soares F, Chea E, Le Bourhis L. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat Immunol. 2010;11:55-62. [PubMed] [DOI] [Cited in This Article: ] |
| 29. | Saitoh T, Fujita N, Jang MH, Uematsu S, Yang BG, Satoh T, Omori H, Noda T, Yamamoto N, Komatsu M. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature. 2008;456:264-268. [PubMed] [DOI] [Cited in This Article: ] |
| 30. | Mizushima N, Noda T, Ohsumi Y. Apg16p is required for the function of the Apg12p-Apg5p conjugate in the yeast autophagy pathway. EMBO J. 1999;18:3888-3896. [PubMed] [DOI] [Cited in This Article: ] |
| 31. | Mizushima N, Kuma A, Kobayashi Y, Yamamoto A, Matsubae M, Takao T, Natsume T, Ohsumi Y, Yoshimori T. Mouse Apg16L, a novel WD-repeat protein, targets to the autophagic isolation membrane with the Apg12-Apg5 conjugate. J Cell Sci. 2003;116:1679-1688. [PubMed] [Cited in This Article: ] |
| 32. | Prescott NJ, Fisher SA, Franke A, Hampe J, Onnie CM, Soars D, Bagnall R, Mirza MM, Sanderson J, Forbes A. A nonsynonymous SNP in ATG16L1 predisposes to ileal Crohn’s disease and is independent of CARD15 and IBD5. Gastroenterology. 2007;132:1665-1671. [PubMed] [DOI] [Cited in This Article: ] |
| 33. | Cheng JF, Ning YJ, Zhang W, Lu ZH, Lin L. T300A polymorphism of ATG16L1 and susceptibility to inflammatory bowel diseases: a meta-analysis. World J Gastroenterol. 2010;16:1258-1266. [PubMed] [DOI] [Cited in This Article: ] |
| 34. | Cadwell K, Liu JY, Brown SL, Miyoshi H, Loh J, Lennerz JK, Kishi C, Kc W, Carrero JA, Hunt S. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 2008;456:259-263. [PubMed] [DOI] [Cited in This Article: ] |
| 35. | Lassen KG, Kuballa P, Conway KL, Patel KK, Becker CE, Peloquin JM, Villablanca EJ, Norman JM, Liu TC, Heath RJ. Atg16L1 T300A variant decreases selective autophagy resulting in altered cytokine signaling and decreased antibacterial defense. Proc Natl Acad Sci USA. 2014;111:7741-7746. [PubMed] [DOI] [Cited in This Article: ] |
| 36. | Murthy A, Li Y, Peng I, Reichelt M, Katakam AK, Noubade R, Roose-Girma M, DeVoss J, Diehl L, Graham RR. A Crohn’s disease variant in Atg16l1 enhances its degradation by caspase 3. Nature. 2014;506:456-462. [PubMed] [DOI] [Cited in This Article: ] |
| 37. | Cadwell K, Patel KK, Maloney NS, Liu TC, Ng AC, Storer CE, Head RD, Xavier R, Stappenbeck TS, Virgin HW. Virus-plus-susceptibility gene interaction determines Crohn’s disease gene Atg16L1 phenotypes in intestine. Cell. 2010;141:1135-1145. [PubMed] [DOI] [Cited in This Article: ] |
| 38. | Chu H, Khosravi A, Kusumawardhani IP, Kwon AH, Vasconcelos AC, Cunha LD, Mayer AE, Shen Y, Wu WL, Kambal A. Gene-microbiota interactions contribute to the pathogenesis of inflammatory bowel disease. Science. 2016;352:1116-1120. [PubMed] [DOI] [Cited in This Article: ] |
| 39. | Parkes M, Barrett JC, Prescott NJ, Tremelling M, Anderson CA, Fisher SA, Roberts RG, Nimmo ER, Cummings FR, Soars D. Sequence variants in the autophagy gene IRGM and multiple other replicating loci contribute to Crohn’s disease susceptibility. Nat Genet. 2007;39:830-832. [PubMed] [DOI] [Cited in This Article: ] |
| 40. | Singh SB, Davis AS, Taylor GA, Deretic V. Human IRGM induces autophagy to eliminate intracellular mycobacteria. Science. 2006;313:1438-1441. [PubMed] [DOI] [Cited in This Article: ] |
| 41. | McCarroll SA, Huett A, Kuballa P, Chilewski SD, Landry A, Goyette P, Zody MC, Hall JL, Brant SR, Cho JH. Deletion polymorphism upstream of IRGM associated with altered IRGM expression and Crohn’s disease. Nat Genet. 2008;40:1107-1112. [PubMed] [DOI] [Cited in This Article: ] |
| 42. | Rufini S, Ciccacci C, Di Fusco D, Ruffa A, Pallone F, Novelli G, Biancone L, Borgiani P. Autophagy and inflammatory bowel disease: Association between variants of the autophagy-related IRGM gene and susceptibility to Crohn’s disease. Dig Liver Dis. 2015;47:744-750. [PubMed] [DOI] [Cited in This Article: ] |
| 43. | Fitch E, Harper E, Skorcheva I, Kurtz SE, Blauvelt A. Pathophysiology of psoriasis: recent advances on IL-23 and Th17 cytokines. Curr Rheumatol Rep. 2007;9:461-467. [PubMed] [Cited in This Article: ] |
| 44. | McGovern D, Powrie F. The IL23 axis plays a key role in the pathogenesis of IBD. Gut. 2007;56:1333-1336. [PubMed] [DOI] [Cited in This Article: ] |
| 45. | Yen D, Cheung J, Scheerens H, Poulet F, McClanahan T, McKenzie B, Kleinschek MA, Owyang A, Mattson J, Blumenschein W. IL-23 is essential for T cell-mediated colitis and promotes inflammation via IL-17 and IL-6. J Clin Invest. 2006;116:1310-1316. [PubMed] [DOI] [Cited in This Article: ] |
| 46. | Parham C, Chirica M, Timans J, Vaisberg E, Travis M, Cheung J, Pflanz S, Zhang R, Singh KP, Vega F. A receptor for the heterodimeric cytokine IL-23 is composed of IL-12Rbeta1 and a novel cytokine receptor subunit, IL-23R. J Immunol. 2002;168:5699-5708. [PubMed] [DOI] [Cited in This Article: ] |
| 47. | Bianco AM, Girardelli M, Tommasini A. Genetics of inflammatory bowel disease from multifactorial to monogenic forms. World J Gastroenterol. 2015;21:12296-12310. [PubMed] [DOI] [Cited in This Article: ] |
| 48. | Fuyuno Y, Yamazaki K, Takahashi A, Esaki M, Kawaguchi T, Takazoe M, Matsumoto T, Matsui T, Tanaka H, Motoya S. Genetic characteristics of inflammatory bowel disease in a Japanese population. J Gastroenterol. 2016;51:672-681. [PubMed] [DOI] [Cited in This Article: ] |
| 49. | Peral de Castro C, Jones SA, Ní Cheallaigh C, Hearnden CA, Williams L, Winter J, Lavelle EC, Mills KH, Harris J. Autophagy regulates IL-23 secretion and innate T cell responses through effects on IL-1 secretion. J Immunol. 2012;189:4144-4153. [PubMed] [DOI] [Cited in This Article: ] |
| 50. | Crook NE, Clem RJ, Miller LK. An apoptosis-inhibiting baculovirus gene with a zinc finger-like motif. J Virol. 1993;67:2168-2174. [PubMed] [Cited in This Article: ] |
| 51. | Eckelman BP, Salvesen GS, Scott FL. Human inhibitor of apoptosis proteins: why XIAP is the black sheep of the family. EMBO Rep. 2006;7:988-994. [PubMed] [DOI] [Cited in This Article: ] |
| 52. | Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069-1075. [PubMed] [DOI] [Cited in This Article: ] |
| 53. | Huang X, Wu Z, Mei Y, Wu M. XIAP inhibits autophagy via XIAP-Mdm2-p53 signalling. EMBO J. 2013;32:2204-2216. [PubMed] [DOI] [Cited in This Article: ] |
| 54. | Zeissig Y, Petersen BS, Milutinovic S, Bosse E, Mayr G, Peuker K, Hartwig J, Keller A, Kohl M, Laass MW. XIAP variants in male Crohn’s disease. Gut. 2015;64:66-76. [PubMed] [DOI] [Cited in This Article: ] |
| 55. | Schwerd T, Pandey S, Yang HT, Bagola K, Jameson E, Jung J, Lachmann RH, Shah N, Patel SY, Booth C. Impaired antibacterial autophagy links granulomatous intestinal inflammation in Niemann-Pick disease type C1 and XIAP deficiency with NOD2 variants in Crohn’s disease. Gut. 2016; Epub ahead of print. [PubMed] [DOI] [Cited in This Article: ] |
| 56. | Alegre-Abarrategui J, Christian H, Lufino MM, Mutihac R, Venda LL, Ansorge O, Wade-Martins R. LRRK2 regulates autophagic activity and localizes to specific membrane microdomains in a novel human genomic reporter cellular model. Hum Mol Genet. 2009;18:4022-4034. [PubMed] [DOI] [Cited in This Article: ] |
| 57. | Barrett JC, Hansoul S, Nicolae DL, Cho JH, Duerr RH, Rioux JD, Brant SR, Silverberg MS, Taylor KD, Barmada MM. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn’s disease. Nat Genet. 2008;40:955-962. [PubMed] [DOI] [Cited in This Article: ] |
| 58. | Gardet A, Benita Y, Li C, Sands BE, Ballester I, Stevens C, Korzenik JR, Rioux JD, Daly MJ, Xavier RJ. LRRK2 is involved in the IFN-gamma response and host response to pathogens. J Immunol. 2010;185:5577-5585. [PubMed] [DOI] [Cited in This Article: ] |
| 59. | Henckaerts L, Cleynen I, Brinar M, John JM, Van Steen K, Rutgeerts P, Vermeire S. Genetic variation in the autophagy gene ULK1 and risk of Crohn’s disease. Inflamm Bowel Dis. 2011;17:1392-1397. [PubMed] [DOI] [Cited in This Article: ] |
| 60. | Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132-141. [PubMed] [DOI] [Cited in This Article: ] |
| 61. | Lee JW, Park S, Takahashi Y, Wang HG. The association of AMPK with ULK1 regulates autophagy. PLoS One. 2010;5:e15394. [PubMed] [DOI] [Cited in This Article: ] |
| 62. | Pei FH, Wang YJ, Gao SL, Liu BR, DU YJ, Liu W, Yu HY, Zhao LX, Chi BR. Vitamin D receptor gene polymorphism and ulcerative colitis susceptibility in Han Chinese. J Dig Dis. 2011;12:90-98. [PubMed] [DOI] [Cited in This Article: ] |
| 63. | Simmons JD, Mullighan C, Welsh KI, Jewell DP. Vitamin D receptor gene polymorphism: association with Crohn’s disease susceptibility. Gut. 2000;47:211-214. [PubMed] [Cited in This Article: ] |
| 64. | Wu S, Zhang YG, Lu R, Xia Y, Zhou D, Petrof EO, Claud EC, Chen D, Chang EB, Carmeliet G. Intestinal epithelial vitamin D receptor deletion leads to defective autophagy in colitis. Gut. 2015;64:1082-1094. [PubMed] [DOI] [Cited in This Article: ] |
| 65. | Abreu-Delgado Y, Isidro RA, Torres EA, González A, Cruz ML, Isidro AA, González-Keelan CI, Medero P, Appleyard CB. Serum vitamin D and colonic vitamin D receptor in inflammatory bowel disease. World J Gastroenterol. 2016;22:3581-3591. [PubMed] [DOI] [Cited in This Article: ] |
| 66. | Henderson P, van Limbergen JE, Wilson DC, Satsangi J, Russell RK. Genetics of childhood-onset inflammatory bowel disease. Inflamm Bowel Dis. 2011;17:346-361. [PubMed] [DOI] [Cited in This Article: ] |
| 67. | Franke A, McGovern DP, Barrett JC, Wang K, Radford-Smith GL, Ahmad T, Lees CW, Balschun T, Lee J, Roberts R. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat Genet. 2010;42:1118-1125. [PubMed] [DOI] [Cited in This Article: ] |
| 68. | Lahiri A, Hedl M, Abraham C. MTMR3 risk allele enhances innate receptor-induced signaling and cytokines by decreasing autophagy and increasing caspase-1 activation. Proc Natl Acad Sci USA. 2015;112:10461-10466. [PubMed] [DOI] [Cited in This Article: ] |
| 69. | Yamamoto-Furusho JK, Peñaloza-Coronel A, Sánchez-Muñoz F, Barreto-Zuñiga R, Dominguez-Lopez A. Peroxisome proliferator-activated receptor-gamma (PPAR-γ) expression is downregulated in patients with active ulcerative colitis. Inflamm Bowel Dis. 2011;17:680-681. [PubMed] [DOI] [Cited in This Article: ] |
| 70. | Rousseaux C, Lefebvre B, Dubuquoy L, Lefebvre P, Romano O, Auwerx J, Metzger D, Wahli W, Desvergne B, Naccari GC. Intestinal antiinflammatory effect of 5-aminosalicylic acid is dependent on peroxisome proliferator-activated receptor-gamma. J Exp Med. 2005;201:1205-1215. [PubMed] [DOI] [Cited in This Article: ] |
| 71. | Chacon-Cabrera A, Fermoselle C, Urtreger AJ, Mateu-Jimenez M, Diament MJ, de Kier Joffé ED, Sandri M, Barreiro E. Pharmacological strategies in lung cancer-induced cachexia: effects on muscle proteolysis, autophagy, structure, and weakness. J Cell Physiol. 2014;229:1660-1672. [PubMed] [DOI] [Cited in This Article: ] |
| 72. | Kuenzig ME, Rezaie A, Seow CH, Otley AR, Steinhart AH, Griffiths AM, Kaplan GG, Benchimol EI. Budesonide for maintenance of remission in Crohn’s disease. Cochrane Database Syst Rev. 2014;CD002913. [PubMed] [DOI] [Cited in This Article: ] |
| 73. | Polman JA, Hunter RG, Speksnijder N, van den Oever JM, Korobko OB, McEwen BS, de Kloet ER, Datson NA. Glucocorticoids modulate the mTOR pathway in the hippocampus: differential effects depending on stress history. Endocrinology. 2012;153:4317-4327. [PubMed] [DOI] [Cited in This Article: ] |
| 74. | Wang H, Kubica N, Ellisen LW, Jefferson LS, Kimball SR. Dexamethasone represses signaling through the mammalian target of rapamycin in muscle cells by enhancing expression of REDD1. J Biol Chem. 2006;281:39128-39134. [PubMed] [DOI] [Cited in This Article: ] |
| 75. | Swerdlow S, McColl K, Rong Y, Lam M, Gupta A, Distelhorst CW. Apoptosis inhibition by Bcl-2 gives way to autophagy in glucocorticoid-treated lymphocytes. Autophagy. 2008;4:612-620. [PubMed] [Cited in This Article: ] |
| 76. | Jia G, Cheng G, Gangahar DM, Agrawal DK. Insulin-like growth factor-1 and TNF-alpha regulate autophagy through c-jun N-terminal kinase and Akt pathways in human atherosclerotic vascular smooth cells. Immunol Cell Biol. 2006;84:448-454. [PubMed] [DOI] [Cited in This Article: ] |
| 77. | Xia X, Kar R, Gluhak-Heinrich J, Yao W, Lane NE, Bonewald LF, Biswas SK, Lo WK, Jiang JX. Glucocorticoid-induced autophagy in osteocytes. J Bone Miner Res. 2010;25:2479-2488. [PubMed] [DOI] [Cited in This Article: ] |
| 78. | Guijarro LG, Román ID, Fernández-Moreno MD, Gisbert JP, Hernández-Breijo B. Is the autophagy induced by thiopurines beneficial or deleterious? Curr Drug Metab. 2012;13:1267-1276. [PubMed] [Cited in This Article: ] |
| 79. | Wang L, Weinshilboum R. Thiopurine S-methyltransferase pharmacogenetics: insights, challenges and future directions. Oncogene. 2006;25:1629-1638. [PubMed] [DOI] [Cited in This Article: ] |
| 80. | Evans WE, Horner M, Chu YQ, Kalwinsky D, Roberts WM. Altered mercaptopurine metabolism, toxic effects, and dosage requirement in a thiopurine methyltransferase-deficient child with acute lymphocytic leukemia. J Pediatr. 1991;119:985-989. [PubMed] [Cited in This Article: ] |
| 81. | Lennard L, Van Loon JA, Weinshilboum RM. Pharmacogenetics of acute azathioprine toxicity: relationship to thiopurine methyltransferase genetic polymorphism. Clin Pharmacol Ther. 1989;46:149-154. [PubMed] [Cited in This Article: ] |
| 82. | Schaeffeler E, Fischer C, Brockmeier D, Wernet D, Moerike K, Eichelbaum M, Zanger UM, Schwab M. Comprehensive analysis of thiopurine S-methyltransferase phenotype-genotype correlation in a large population of German-Caucasians and identification of novel TPMT variants. Pharmacogenetics. 2004;14:407-417. [PubMed] [Cited in This Article: ] |
| 83. | Schütz E, Gummert J, Mohr F, Oellerich M. Azathioprine-induced myelosuppression in thiopurine methyltransferase deficient heart transplant recipient. Lancet. 1993;341:436. [PubMed] [Cited in This Article: ] |
| 84. | Li F, Wang L, Burgess RJ, Weinshilboum RM. Thiopurine S-methyltransferase pharmacogenetics: autophagy as a mechanism for variant allozyme degradation. Pharmacogenet Genomics. 2008;18:1083-1094. [PubMed] [DOI] [Cited in This Article: ] |
| 85. | Markowitz J, Grancher K, Kohn N, Daum F. Immunomodulatory therapy for pediatric inflammatory bowel disease: changing patterns of use, 1990-2000. Am J Gastroenterol. 2002;97:928-932. [PubMed] [DOI] [Cited in This Article: ] |
| 86. | Nuki Y, Esaki M, Asano K, Maehata Y, Umeno J, Moriyama T, Nakamura S, Matsumoto T, Kitazono T. Comparison of the therapeutic efficacy and safety between tacrolimus and infliximab for moderate-to-severe ulcerative colitis: a single center experience. Scand J Gastroenterol. 2016;51:700-705. [PubMed] [DOI] [Cited in This Article: ] |
| 87. | Pallet N, Bouvier N, Legendre C, Gilleron J, Codogno P, Beaune P, Thervet E, Anglicheau D. Autophagy protects renal tubular cells against cyclosporine toxicity. Autophagy. 2008;4:783-791. [PubMed] [Cited in This Article: ] |
| 88. | Kimura T, Takahashi A, Takabatake Y, Namba T, Yamamoto T, Kaimori JY, Matsui I, Kitamura H, Niimura F, Matsusaka T. Autophagy protects kidney proximal tubule epithelial cells from mitochondrial metabolic stress. Autophagy. 2013;9:1876-1886. [PubMed] [DOI] [Cited in This Article: ] |
| 89. | Kim HS, Choi SI, Jeung EB, Yoo YM. Cyclosporine A induces apoptotic and autophagic cell death in rat pituitary GH3 cells. PLoS One. 2014;9:e108981. [PubMed] [DOI] [Cited in This Article: ] |
| 90. | Liu J, Albers MW, Wandless TJ, Luan S, Alberg DG, Belshaw PJ, Cohen P, MacKintosh C, Klee CB, Schreiber SL. Inhibition of T cell signaling by immunophilin-ligand complexes correlates with loss of calcineurin phosphatase activity. Biochemistry. 1992;31:3896-3901. [PubMed] [Cited in This Article: ] |
| 91. | Jung CH, Jun CB, Ro SH, Kim YM, Otto NM, Cao J, Kundu M, Kim DH. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell. 2009;20:1992-2003. [PubMed] [DOI] [Cited in This Article: ] |
| 92. | Ganley IG, Lam du H, Wang J, Ding X, Chen S, Jiang X. ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J Biol Chem. 2009;284:12297-12305. [PubMed] [DOI] [Cited in This Article: ] |
| 93. | de Ridder L, Waterman M, Turner D, Bronsky J, Hauer AC, Dias JA, Strisciuglio C, Ruemmele FM, Levine A, Lionetti P. Use of Biosimilars in Paediatric Inflammatory Bowel Disease: A Position Statement of the ESPGHAN Paediatric IBD Porto Group. J Pediatr Gastroenterol Nutr. 2015;61:503-508. [PubMed] [DOI] [Cited in This Article: ] |
| 94. | Connor AM, Mahomed N, Gandhi R, Keystone EC, Berger SA. TNFα modulates protein degradation pathways in rheumatoid arthritis synovial fibroblasts. Arthritis Res Ther. 2012;14:R62. [PubMed] [DOI] [Cited in This Article: ] |
| 95. | Keller CW, Fokken C, Turville SG, Lünemann A, Schmidt J, Münz C, Lünemann JD. TNF-alpha induces macroautophagy and regulates MHC class II expression in human skeletal muscle cells. J Biol Chem. 2011;286:3970-3980. [PubMed] [DOI] [Cited in This Article: ] |
| 96. | Cha HH, Hwang JR, Kim HY, Choi SJ, Oh SY, Roh CR. Autophagy induced by tumor necrosis factor α mediates intrinsic apoptosis in trophoblastic cells. Reprod Sci. 2014;21:612-622. [PubMed] [DOI] [Cited in This Article: ] |
| 97. | Nys K, Agostinis P, Vermeire S. Autophagy: a new target or an old strategy for the treatment of Crohn’s disease? Nat Rev Gastroenterol Hepatol. 2013;10:395-401. [PubMed] [DOI] [Cited in This Article: ] |