Copyright
©The Author(s) 2016.
World J Gastroenterol. Jul 14, 2016; 22(26): 6016-6026
Published online Jul 14, 2016. doi: 10.3748/wjg.v22.i26.6016
Published online Jul 14, 2016. doi: 10.3748/wjg.v22.i26.6016
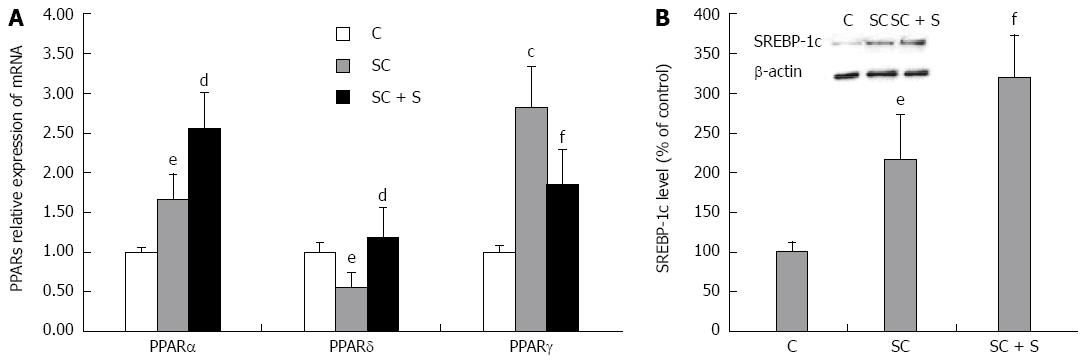
Figure 2 Effects of silybin on transcription factors regulating lipid metabolism.
In control (C) and steatotic FaO cells incubated in the absence (SC) or in the presence (SC + S) of silybin 50 μmol/L we assessed. A: mRNA expression of PPARα, PPARδ and PPARγ by qPCR; GAPDH was used as the internal control for quantifying gene expression; data expressed as fold induction with respect to controls; B: Densitometric analysis of SREBP-1c by Western blotting; β-actin was the protein loading control used as housekeeping gene for normalization; data are expressed as percentage values with respect to controls. Values are mean ± SD from at least three independent experiments. ANOVA followed by Tukey’s test was used to assess the statistical significance between groups Significant differences are denoted by symbols: cP≤ 0.01, eP≤ 0.05 C vs OP and dP≤ 0.01, fP≤ 0.05 OP vs Slybin.
- Citation: Vecchione G, Grasselli E, Voci A, Baldini F, Grattagliano I, Wang DQ, Portincasa P, Vergani L. Silybin counteracts lipid excess and oxidative stress in cultured steatotic hepatic cells. World J Gastroenterol 2016; 22(26): 6016-6026
- URL: https://www.wjgnet.com/1007-9327/full/v22/i26/6016.htm
- DOI: https://dx.doi.org/10.3748/wjg.v22.i26.6016