Copyright
©The Author(s) 2015.
World J Gastroenterol. Dec 21, 2015; 21(47): 13360-13367
Published online Dec 21, 2015. doi: 10.3748/wjg.v21.i47.13360
Published online Dec 21, 2015. doi: 10.3748/wjg.v21.i47.13360
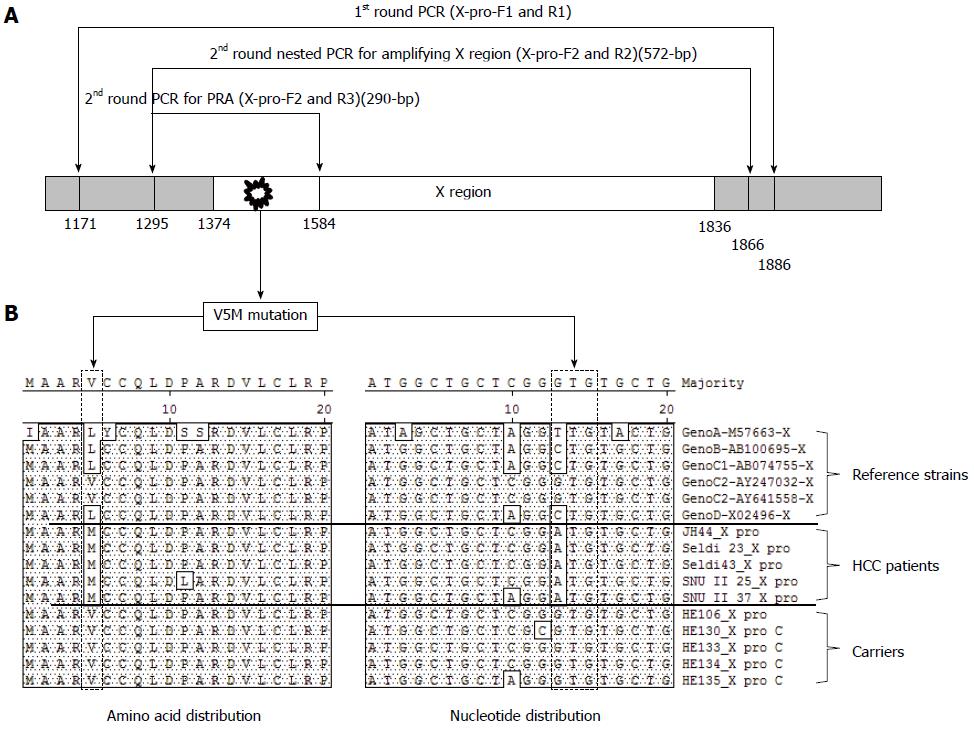
Figure 1 Polymerase chain reaction amplification and direct sequencing analysis of hepatitis B virus X region.
A: Location of primer set for the nested PCR direct sequencing (1st primer set; X-pro-F1 and X-pro-R1, 2nd primer set; X-pro-F2 and X-pro-R2) and Fok-I based nested PRA (1st primer set; X-pro-F1 and X-pro-R1, 2nd primer set; X-pro-F2 and X-pro-R3) for the detection of HBxAg V5M mutations and the 3 polymorphisms [V5(GTG), M5(ATG), and L5(CTG or TTG)] of the 5th codon of HBxAg; B: The amino acid and nucleotide sequences of the X region from six reference strains (Genotype A, B, C1, C2, and D) and 10 chronic hepatitis patients (five carriers and five HCC patients) were aligned.
-
Citation: Kim H, Hong SH, Lee SA, Gong JR, Kim BJ. Development of
Fok -I based nested polymerase chain reaction-restriction fragment length polymorphism analysis for detection of hepatitis B virus X region V5M mutation. World J Gastroenterol 2015; 21(47): 13360-13367 - URL: https://www.wjgnet.com/1007-9327/full/v21/i47/13360.htm
- DOI: https://dx.doi.org/10.3748/wjg.v21.i47.13360