Copyright
©The Author(s) 2015.
World J Gastroenterol. Jul 28, 2015; 21(28): 8492-8507
Published online Jul 28, 2015. doi: 10.3748/wjg.v21.i28.8492
Published online Jul 28, 2015. doi: 10.3748/wjg.v21.i28.8492
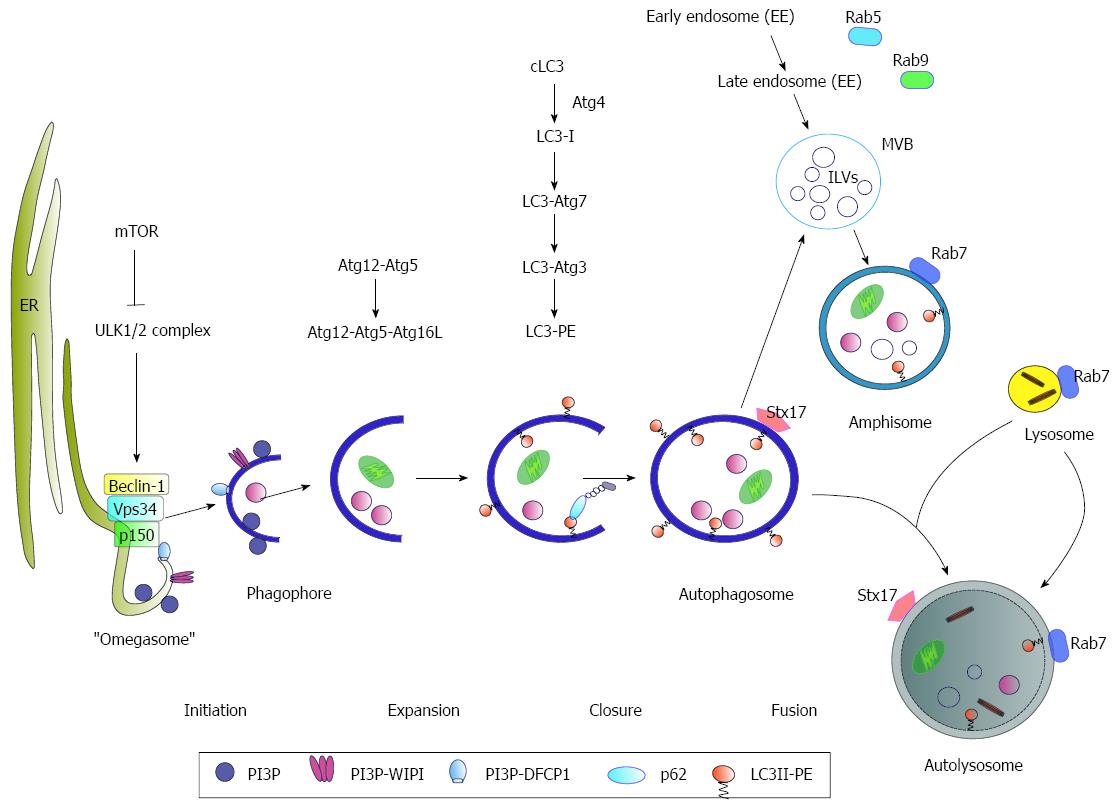
Figure 1 Schematic model of autophagy.
Under nutrient-rich conditions mammalian target of rapamycin (mTOR) represses the ULK1/2 complex. During starvation, mTOR kinase is inhibited resulting in activation of autophagy. As an initial step, Beclin-1 forms a complex with Vps34 and p150 (PI3K complex). The activated ULK1/2 and PI3K complex catalyze the formation of a PI3P (phosphatidylinositol-3-phosphate)-enriched environment. The PI3P further recruits the effector double FYVE-containing protein 1 (DFCP1) and WD-repeat domain phosphoinositide-interacting (WIPI) proteins, leading to phagophore nucleation by formation of an ER-associated Ω-structure (“omegasome”). After the nucleation process the formation of the autophagosome occurs. Here, the Atg5-Atg12-Atg16L complex and LC3 II, conjugated to phosphatidylethanolamine (PE) (LC3-PE), play a crucial role in the elongation and closure of the membrane. The autophagosomes can either directly fuse with lysosomes to form the autolysosome or fuse with late endosomes/multivesicular bodies (MVBs) forming an amphisome prior fusion with the lysosome where their cargo is digested. Rab7-GTPase activity triggers the fusion of the autophagosome with the lysosome. Another factor involved in autophagosome-lysosome fusion is the autophagosomal SNARE (soluble N-ethylmaleimide-sensitive factor attachment protein) syntaxin 17 (Stx17) that can be found on the outer membrane of completed enclosed autophagosomes.
- Citation: Ploen D, Hildt E. Hepatitis C virus comes for dinner: How the hepatitis C virus interferes with autophagy. World J Gastroenterol 2015; 21(28): 8492-8507
- URL: https://www.wjgnet.com/1007-9327/full/v21/i28/8492.htm
- DOI: https://dx.doi.org/10.3748/wjg.v21.i28.8492