Copyright
©2014 Baishideng Publishing Group Inc.
World J Gastroenterol. Dec 14, 2014; 20(46): 17376-17387
Published online Dec 14, 2014. doi: 10.3748/wjg.v20.i46.17376
Published online Dec 14, 2014. doi: 10.3748/wjg.v20.i46.17376
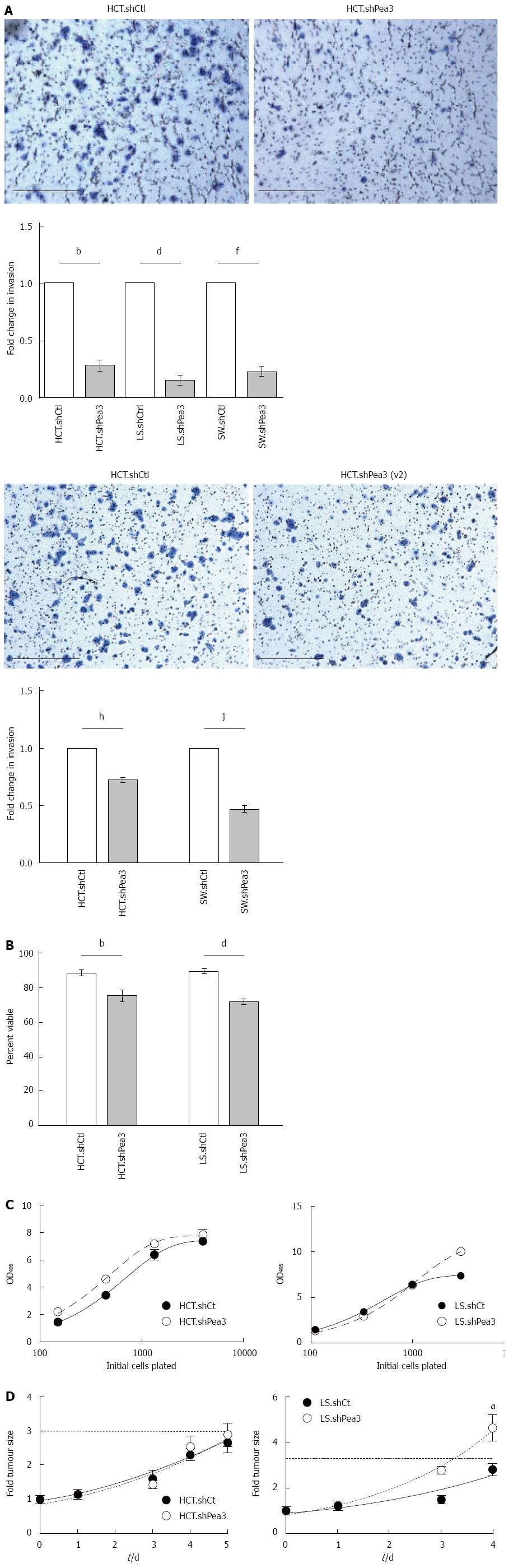
Figure 3 Characterization of invasiveness, anoikis resistance, and proliferative capacity of colorectal carcinoma cells expressing the short hairpin RNA Pea3 (shPea3) construct.
A: Representative images of the 24-h transwell invasion assay using HCT.shPea3, HCT.shPea3(v2) and HCT.shCtrl cells with quantification of the transwell invasion assays. Data are shown as fold change in invasion of shPea3 or shPea3(v2) expressing cells relative to their shCtrl counterparts. bP < 0.01, HCT.shCtrl vs HCT.shPea3; dP < 0.01, LS.shCtrl vs LS.shPea3; fP < 0.01, SW.shCtrl vs SW.shPea3; hP < 0.01, LS.shCtrl vs LS.shPea3; B: Anoikis assay using the stable HCT116 and LS174T transductants. Cell death was assessed using dye exclusion, and the fraction of surviving cells after 2 d (HCT116) or 5 d (LS174T) was plotted. bP < 0.01, HCT.shCtrl vs HCT.shPea3; dP < 0.01, LS.shCtrl vs LS.shPea3; C: In vitro proliferation assay of HCT.shPea3 and HCT.shCtrl cells (left panel), as well as LS.shPea3 and LS.shCtrl cells (right panel). Cells were plated in 96-wells in 4-fold dilution series, followed by OD reading at 495 nm for quantification (shown). Area under curve analysis was performed for statistical significance; D: In vivo tumour xenograft growth assay of HCT.shPea3 and LS.shPea3 cells. 5 × 106 cells were injected s.c. into the flanks of mice (8 per group) and calliper measurements of the tumours were taken. Shown are fold change in tumour sizes compared to day zero. Dotted horizontal line indicates 3-fold starting tumour size. aP < 0.05 vs final tumour measurements. Statistical analysis was performed using Student’s t-test for the final tumour measurements.
- Citation: Mesci A, Taeb S, Huang X, Jairath R, Sivaloganathan D, Liu SK. Pea3 expression promotes the invasive and metastatic potential of colorectal carcinoma. World J Gastroenterol 2014; 20(46): 17376-17387
- URL: https://www.wjgnet.com/1007-9327/full/v20/i46/17376.htm
- DOI: https://dx.doi.org/10.3748/wjg.v20.i46.17376