INTRODUCTION
Hepatocellular carcinoma (HCC) is the fifth most common malignancy worldwide, and > 80% of HCC occurs in developing countries[1]. HCC remains a significant concern of cancer research because of its poor survival rate and high rates of recurrence[2,3]. Prognosis of surgical or loco-regional therapies for patients with intermediate- and advanced-stage disease remains poor[4]. Several effective gene-targeting agents are currently being tested in preclinical studies[5-9]. Unfortunately, no chemotherapy is effective for HCC patients.
Kladney et al[10] first isolated Golgi protein (GP)73 in a genetic screen. Expression of GP73 is increased markedly in HCC cells and its serum levels appear to be predictive of HCC[11-15], and several studies have reported the use of GP73 as a serum marker for HCC[16-22]. GP73 may be elevated even when small undetectable tumors are present[23]. The physiological and pathological roles of GP73 have attracted considerable attention in recent years[24-30]. However the function of GP73 in hepatic carcinoma cells remains obscure. The expression of GP73 was silenced in the HCC cell line Bel7402 and Hep G2 by stealth RNAi, which serves as a powerful technology to block specifically the expression of target genes in the present study[31-35]. The effects of GP73 on cell proliferation and apoptosis were also evaluated in this study.
MATERIALS AND METHODS
Stealth RNAi
According to the siRNA design guidelines[27,28], one RNAi target sequence was selected corresponding to the nucleotides of RNAI-Stealth RNAi of the human GP73 mRNA (GenBank Accession No. NM177937.2). The sequence of the synthesized oligonucleotide was: HSS181966: sense 5’-GGAAACGGGCGUCGCAGCAUGAAGU-3’, anti-sense 5’-ACUUCAUGC UGCUACGCCCGUUUCC-3’.
Transfection
Lipofectamine RNAi Max transfection agent (Invitrogen, Carlsbad, CA, United States) was used to transfect synthesized Stealth RNAi against GP73 into Hep G2 and Bel7402 cells. BLOCK-iT Alexa Fluor Red Fluorescent (Invitrogen) was used to confirm the transfection efficiency of each duplex siRNA. Stealth RNAi, fluorescent logo or negative control duplexes were delivered into Hep G2 and Bel7402 cells through reverse transfection.
Reverse transcriptase polymerase chain reaction
To analyze quantitatively the effects of Stealth RNAi on GP73 mRNA, cells were transfected with Stealth RNAi or negative control in culture flasks. After 24 and 48 h, cells were harvested by trypsinization and rinsed twice with cold PBS. TRIzol reagent (Invitrogen) was used to extract total RNA. Two-step real-time reverse transcriptase polymerase chain reaction (RT-PCR) kits (TakaRa, Japan) was used to perform first-strand cDNA synthesis and amplification. The 7000 Sequence Detection System (Applied Biosystems, Foster City, CA, United States) was used to perform quantitative PCR amplifications. A 20-μL reaction volume containing 10 μL 2× SYBR Premix Ex TaqTM (TakaRa) was used to carry out this reaction. β-Actin was used as an internal standard. The primer sequences were: GP73 sense 5’-GTGCTGGTGCCAGCCTGTTA-3’ and anti-sense 5’-AGTGCTCTAGGCCA TTGATTGATTG-3’, β-actin sense 5’-GCAAGCAGGAGTATGACGAGT-3’ and anti-sense 5’-GCAAGCAGGAGTATGACGAGT-3’. Thermal cycle conditions: 95 °C for 30 s, followed by 40 cycles of 94 °C for 5 s, and 61 °C for 30 s. The ΔCt of each group was calculated by the formula: ΔCt = CtGP73 - Ctβ-actin. ΔΔCt was calculated by ΔCttreated - ΔCtcontrol. The fold change for GP73 expression levels of the treated groups were calculated using 2-ΔΔCt. The primers described above and the comparative threshold (Ct) method were used to calculate the relative amount of mRNA in the treated samples compared to the control samples. The real-time PCR assays were performed in triplicate.
GP73 proteins detection
To analyze quantitatively the effects of Stealth RNAi on GP73 protein levels in supernatant of Hep G2 and Bel7402 cells, cells were transfected with Stealth RNAi or negative control in culture flasks. After 24 and 48 h, the supernatant was collected, and GP73 protein levels in supernatant of Hep G2 and Bel7402 cells were detected using a commercially available human GP73 ELISA kit. The ELISAs were performed in triplicate.
Cell proliferation assay and cell cycle analysis
Cell proliferation was measured by MTT assay, according to the manufacturer’s instructions. Propidium iodide (PI) staining of the nuclei was used to monitor cell cycle assay. Seventy-five percent cold alcohol was used overnight to fix the cells, and then the cells were resuspended in 300 μL PBS and stained with 500 μL PI (250 μg/mL) for 30 min in the dark. Flow cytometry was used to analyze cells. Cell cycle assays were performed in triplicate.
Apoptosis assessment with flow cytometry and transmission electron microscopy
To analyze quantitatively the effects of Stealth RNAi on apoptosis, cells were transfected with Stealth RNAi or negative control in culture flasks. After 48 h, cells were harvested by trypsinization and rinsed twice with cold PBS. Cells were resuspended in 200 μL binding buffer and then treated with 10 μL Annexin V-FITC and 5 μL PI (Sigma, St Louis, MO, United States) for 15 min. Flow cytometric analysis of cells was performed with an EpicsXL Coulter flow cytometer (Beckman-Coulter, United States). All assays were repeated three times. Transmission electron microscopy (TEM) was used to detect apoptosis. Cells were harvested by trypsinization and rinsed twice with cold PBS. Glutaraldehyde (0.3%) was used to prefix the cells overnight, and 10 mL/L osmic acid was used to post-fix the cells. The cells were observed under a JEM-1230 transmission electron microscope (Jeol, Japan).
Protein extraction and western immunoblot analysis
To analyze quantitatively the mechanism of Stealth RNAi on apoptosis, Hep G2 cells were transfected with Stealth RNAi or negative control in culture flasks. After 48 h, cells were harvested by trypsinization and rinsed twice with cold PBS. Hep G2 cells (5 × 105) were lysed by lysis buffer (phenylmethylsulfonyl fluoride), then drawing the protein standard curve to calculate the density of total protein. Ten percent SDS-PAGE was used to separate proteins, and proteins were transferred to nitrocellulose membranes. Anti-GP73, Bax, Bcl-2, cytochrome c and procaspase-3, β-actin primary antibody (1:1000; Abcam, Cambridge, MA, United States) were used to incubate membranes, and then, anti-rabbit secondary antibody conjugated with horseradish peroxidase was used to incubate membranes (1:5000) again. Western Blotting Substrate (Bio-Rad) was used to visualize immunoreactive proteins. All assays were repeated three times.
Statistical analysis
Mean ± SD was used to express the data. The data among the three groups were compared by one-way analysis of variance followed by Bonferroni correction. Some data were also analyzed by Student’s t test. SPSS for Windows version 19.0 was used for statistical analysis. P < 0.01 was considered significant.
RESULTS
Transfection efficiency
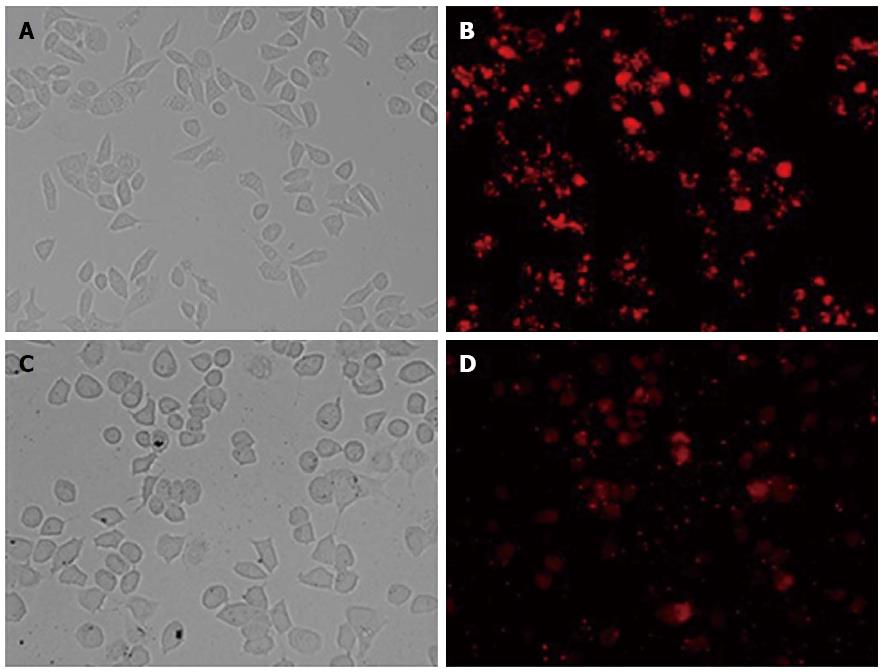
Twenty-four hours after transfection, Bel7402 and Hep G2 cells were observed by fluorescent microscopy. The efficiency of transfection was > 80% when using 30 nmol/L final concentrations of oligo duplex and 1 × 105/mL cells (Figure 1). Therefore, 30 nmol/L siRNA and 1 × 105/mL Bel7402 and Hep G2 cells were used to perform subsequent experiments.
Figure 1 BLOCK-iT Alexa Fluor red fluorescent control.
The BLOCK-iT Alexa Fluor red fluorescent control (30 nmol/L) was transfected into Hep G2 and Bel7402 cells using Lipofectamine RNAi MAX Transfection Reagent. Twenty-four hours after transfection, growth medium was removed and replaced with PBS. Hep G2 and Bel7402 cell localization of the Alexa Fluor Red Fluorescent control oligo is seen with fluorescent microscopy (B and D, × 200). Over 80% of the cells took up the control oligo and retained a normal morphology, as seen in bright field (A and C, × 200).
Expression of GP73 after transfection
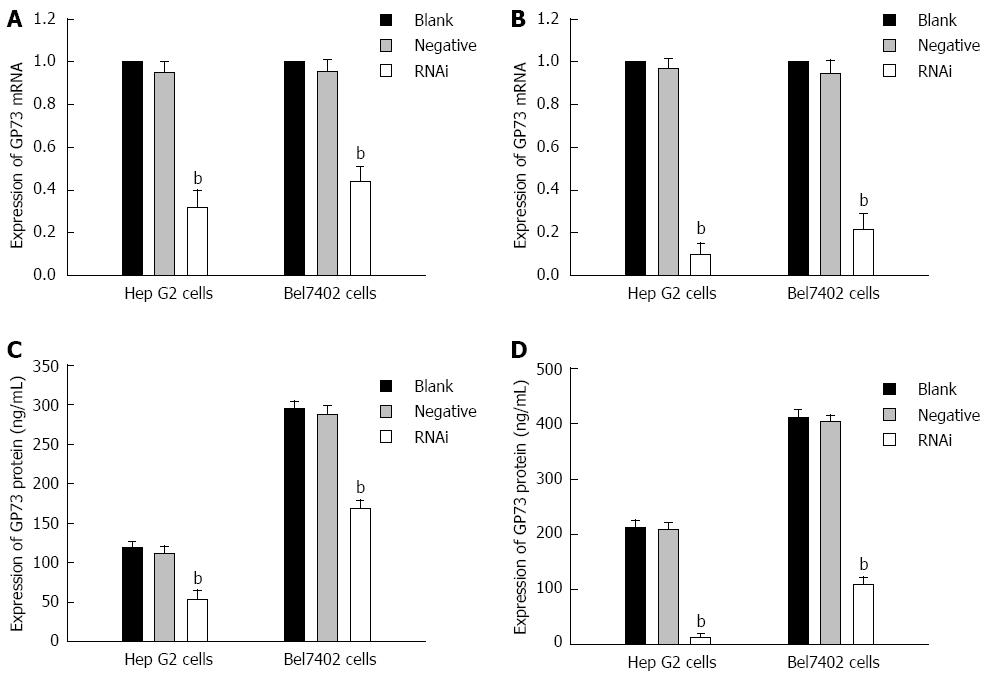
After Stealth siRNA targeting GP73 was transfected into Hep G2 and Bel7402 cells, mRNA levels were measured by RT-PCR. There was an obvious reduction of GP73 mRNA levels in the Stealth RNAi group. Quantification analysis revealed that GP73 mRNA was reduced by 68.7% and 90.3% of the blank control for Hep G2 cells, and 56.7% and 88.7% of the blank control for Bel7402 cells at 24 and 48 h after transfection, respectively. A significant difference was found between the blank control and Stealth RNAi groups (Figure 2A and B, P < 0.01). ELISA of supernatant demonstrated that GP73 protein levels were also significantly reduced compared with the blank control. GP73 protein levels for blank control, negative control and RNAi groups at 24 and 48 h after transfection are shown in Figure 2C and D (P < 0.01). There was a significant difference between the blank control and RNAi groups (Figure 2C and D, P < 0.01). Expression of GP73 in Hep G2 cells was also significantly lower than in Bel7402 cells after transfection.
Figure 2 Expression of GP73 mRNA and protein for Hep G2 and Bel7402 cells after transfection with Stealth RNAi against GP73, negative control RNAi or blank control.
A: Relative expression of GP73 mRNA level at 24 h after transfection analyzed by quantitative real-time reverse transcription polymerase chain reaction (RT-PCR); B: Relative expression of GP73 mRNA level at 48 h after transfection analyzed by quantitative RT-PCR; C: Expression of GP73 proteins in supernatant of Hep G2 and Bel7402 cells were detected by enzyme-linked immunosorbent assay (ELISA) at 24 and 48 h; D: Expression of GP73 proteins in supernatant of Hep G2 and Bel7402 cells was detected by ELISA at 48 h. The corresponding values of GP73 protein in the three groups at 24 and 48 h after transfection. bP < 0.01 vs blank group.
Silencing of GP73 gene decreased viability and proliferation in Bel7402 and Hep G2 cells
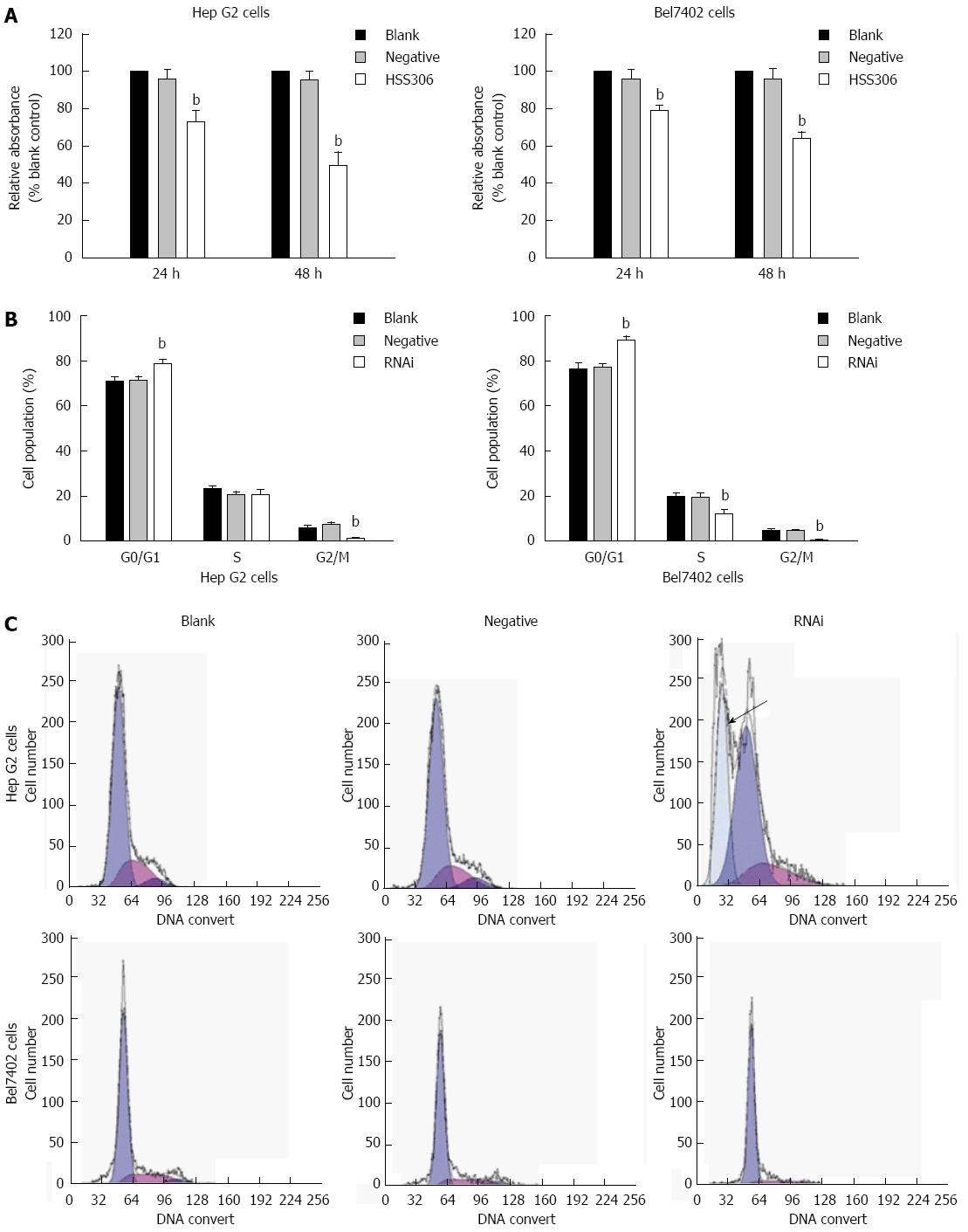
As illustrated in Figure 3A, the viability of Hep G2 and Bel7402 cells was reduced significantly after transfection. The inhibition rate in the RNAi group was 27.09% and 50.53% at 24 and 48 h after transfection for Hep G2 cells, respectively, and 21.3% and 46.4% at 24 and 48 h after transfection for Bel7402 cells (Figure 3A, P < 0.01). As illustrated in Figure 3B and C, an apoptotic peak was seen in the RNAi group of Hep G2 cells, and cell cycle analysis showed that proliferation of Hep G2 cells yielded 78.22% ± 0.35% cells in the G0/G1 phase, 20.72% ± 1.19% cells in S phase, and 1.07% ± 1.09% cells in G2/M phase 48 h after transfection. The percentage of cells in G0/G1 phase increased significantly, however, the percentage of cells in G2/M phase decreased significantly (P < 0.01). Proliferation control Bel7402 cells yielded 88.81% ± 1.13% cells in G0/G1 phase, 12.1% ± 1.12% cells in S phase, and 0.98% ± 1.08% cells in G2/M phase 48 h after transfection in the RNAi group. The percentage of cells in G0/G1 phase increased significantly, however, the percentage of cells in G2/M phase decreased significantly in both cells.
Figure 3 Silencing of GP73 gene decreased cell proliferation.
A: MTT analysis at 24 and 48 h (% blank control) for Hep G2 and Bel7402 cells; B: Cell cycle assay was analyzed by flow cytometry; C: Flow cytometric analysis of the cell cycle distribution of Hep G2 and Bel7402 cells at 24 h after transfection. Cells were washed, fixed, and stained with propidium iodide, and analyzed using Beckman-Coulter Epics flow cytometer. bP < 0.01 vs blank group.
Silencing of GP73 gene induces apoptosis
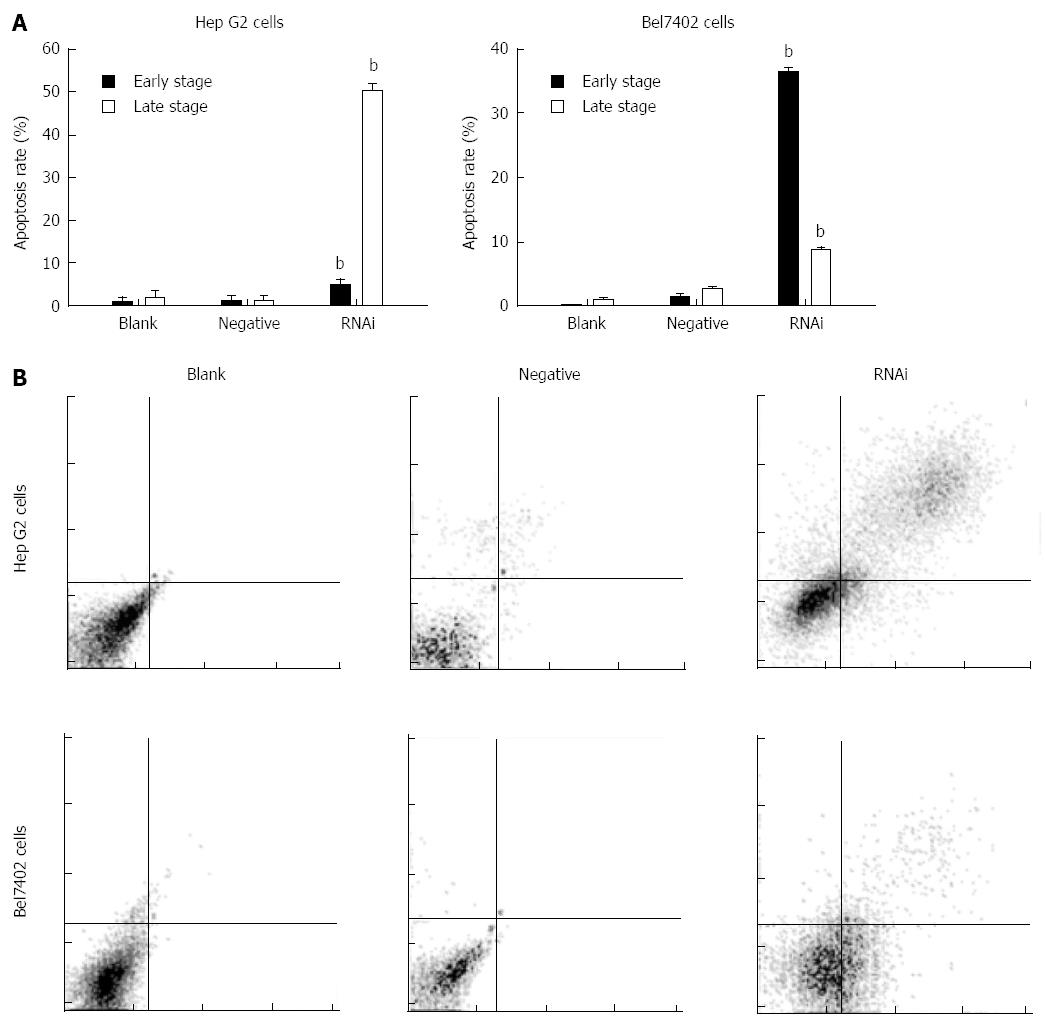
After transfection for 48 h, the apoptotic cells were markedly increased in the RNAi group compared to those in the negative and blank control groups in Hep G2 and Bel7402 cells. Their values are shown in Figure 4 (P < 0.01). The results showed that terminal- and early-stage apoptotic cells were markedly increased in the RNAi group compared to the blank control group in both cells, and terminal-stage apoptosis mainly occurred in Hep G2 cells after transfection, however, early-stage apoptosis mainly occurred in Bel7402 cells.
Figure 4 Effect of GP73-targeted stealth RNAi transfection on apoptosis of Hep G2 and Bel7402 cells.
A: Apoptosis assay was analyzed by flow cytometry; B: Apoptotic effect of Stealth RNAi after 48 h, and later stained with annexin V-FITC and PI. The stained cells were analyzed for apoptosis using a Beckman-Coulter Epics flow cytometer. bP < 0.01 vs blank group.
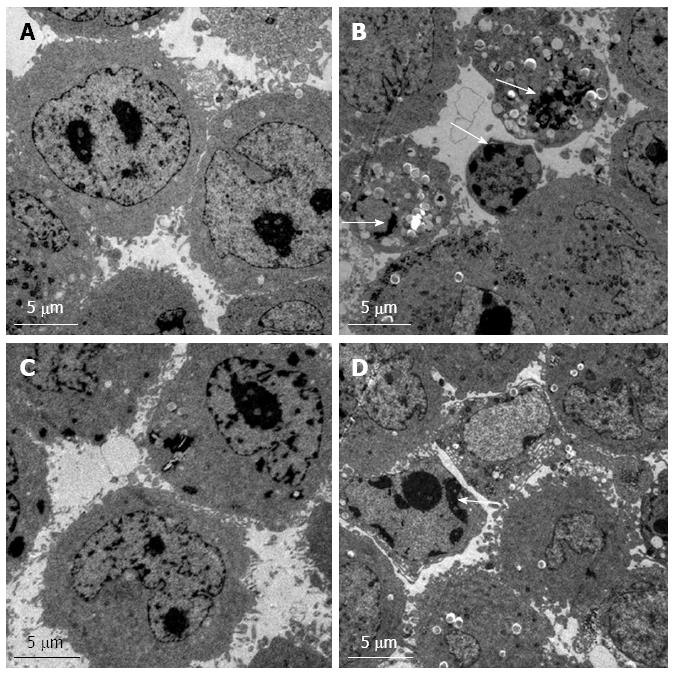
Changes in ultrastructural morphology of cells
Normal control Hep G2 cells were observed by TEM (Figure 5A). Some typical manifestations of apoptosis such as vacuoles, nuclear fragmentation and apoptotic bodies were observed in the RNAi group of Hep G2 cells (Figure 5B). Normal control Bel7402 cells were observed (Figure 5C). Dense chromatin and some vacuoles could be seen in the RNAi group of Bel7402 cells (Figure 5D).
Figure 5 Transmission electron microscopy.
The ultrastructural morphology of control (A) and RNAi (B) of Hep G2 cells. The ultrastructural morphology of control (C) and RNAi (D) Bel7402 cells. Nuclear fragmentation and apoptotic bodies are indicated by arrows.
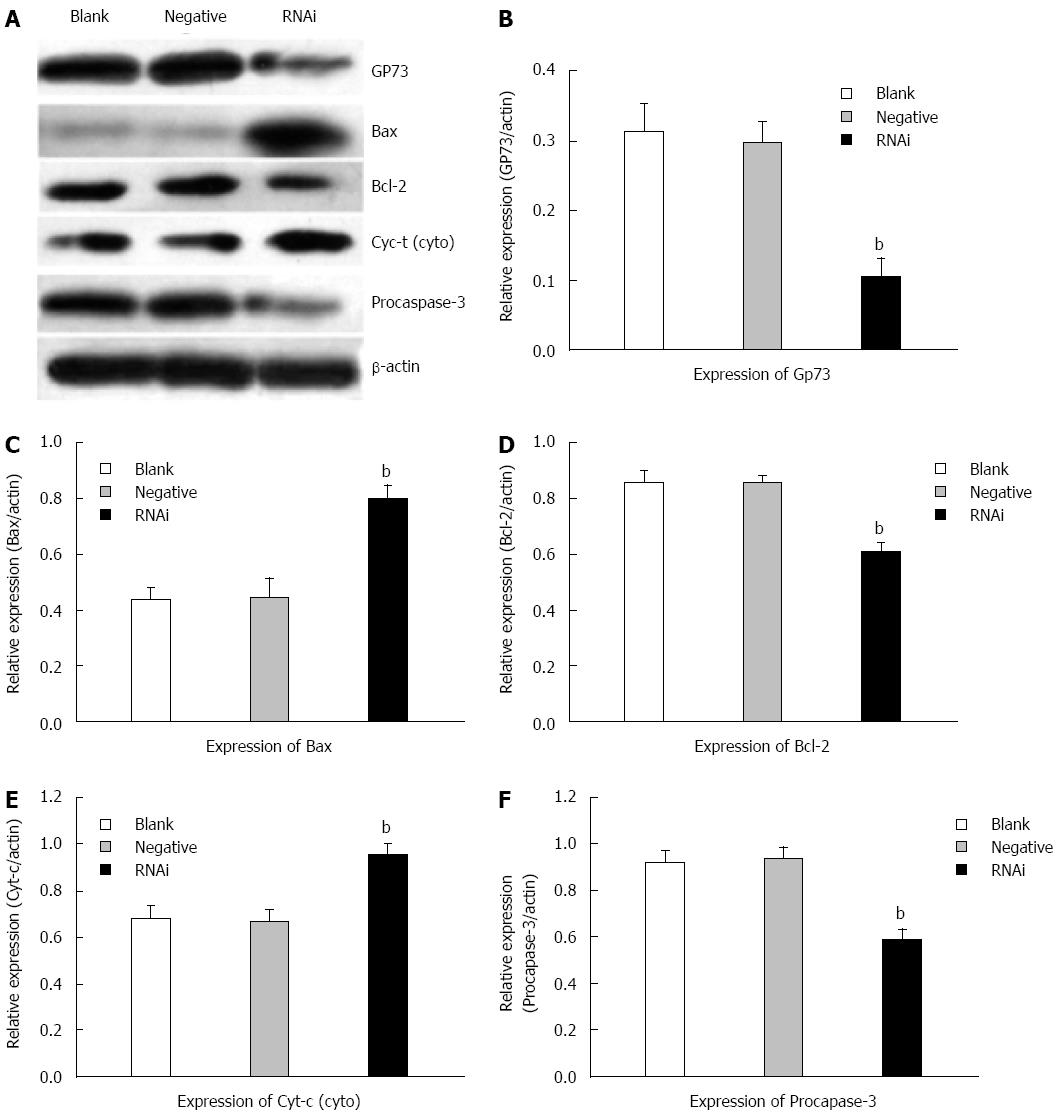
Effects of silencing GP73 gene on apoptosis-related molecules in Hep G2 cells
Western blotting was performed to analyze the expression of GP73, Bax/Bcl-2, cytochrome c and procaspase-3 in Hep G2 cells. The protein levels of GP73 decreased significantly compared with those in negative and blank control groups at 48 h after transfection in Hep G2 cells. Compared with negative and blank control groups, the protein levels of Bcl-2 were reduced significantly, however, the protein levels of Bax were markedly elevated. Compared to the negative and blank control groups at 48 h after transfection, the protein level of cytochrome c in the cytoplasm was markedly elevated in the RNAi group, and the protein level of procaspase-3 was reduced markedly (Figure 6).
Figure 6 Effects of silencing GP73 gene on apoptosis-related molecules in Hep G2 cells.
A: Western blotting detected expression of GP73, Bcl-2, Bax, cytochrome c and procaspase-3 in Hep G2 cells at 48 h after transfection with Stealth RNAi (HSS181966; Stealth RNAi against GP73, negative control RNAi) or blank control; B-F: Representative immunoblots are shown from three independent experiments. Software Glyko Bandscan 5.0 was used to quantify the bands, and the expression of β-actin was used as a loading control, the protein levels were normalized against β-actin, data were expressed as mean ± SD (bP < 0.01 vs blank group).
DISCUSSION
There is a growing body of evidence that GP73 serum levels correlate with the presence of HCC. However, although GP73 levels in the circulation do not appear to be elevated in healthy subjects, these and other reports suggest elevation of the marker in people with inflammatory liver diseases, in the absence of cancer. Thus, its usefulness in distinguishing cancer from cirrhosis or other liver conditions associated with inflammation is still being evaluated. Work by others has found that GP73 levels are affected by tumor necrosis factor-α and interferon-γ in tissue culture systems, and there is an association between levels of GP73 and osmoles in serum from people with liver cirrhosis, but there is no significant correlation in HCC. We do not know the molecular basis for the high expression of GP73 in these cells, and the effect of GP73 in hepatic carcinoma cells is also unclear. We investigated the effect of GP73 in hepatic carcinoma by silencing expression of GP73 through siRNA. In this study, after Stealth RNAi against GP73 was transfected into Hep G2 and Bel7402 cells, the expression of GP73 and mRNA of GP73 decreased significantly in both cells. The decreasing expression of GP73 inhibited cell proliferation and led to apoptosis. Besides, decreasing expression of GP73 also leads to reduction of Bcl-2/Bax ratio, an increase in protein level of cytochrome c in the cytoplasm, and decreasing procaspase-3.
The effects of Stealth RNAi on both GP73 mRNA and protein levels were measured. The results demonstrated that the Stealth RNAi against GP73 (RNAi) can reduce the expression of GP73 effectively at both mRNA and protein levels in Hep G2 and Bel7402 cells after transfection. Western blotting showed that GP73 protein decreased significantly in the RNAi group compared with the other groups. In addition, cell cycle assay confirmed that the proliferation of Bel7402 and Hep G2 cells was reduced significantly after expression of GP73 gene was silenced by RNAi. In Bel7402 and Hep G2 cells, the percentage of cells in G0/G1 phase increased significantly, however, the percentage of cells in G2/M phase decreased significantly. The percentage of cells in S phase decreased in Bel7402 cells after transfection, although the percentage of Hep G2 cells in S phase did not change significantly. However, an apoptotic peak was seen in Hep G2 cells, which indicates that terminal apoptosis had occurred. The proliferation of hepatic cancer cells is related to the expression of GP73. Decreasing expression of GP73 inhibited hepatic cancer cells in G0/G1 phase, and sometimes it also affected hepatic cancer cells in S phase.
Furthermore, fluorescence activated cell sorting methods suggested that Stealth RNAi against GP73 (RNAi) caused accumulation of early-stage apoptotic Bel7402 cells and accumulation of terminal-stage apoptotic Hep G2 cells at 48 h after transfection (GP73 was silenced). The apoptotic peak was also seen in proliferation of Hep G2 cells. The different stage of apoptosis may be related to the different expression of GP73 in Bel7402 and Hep G2 cells. TEM observation agreed with the former results. Dense chromatin appeared near the nuclear membrane, many foaming phenomenonhappened in Bel7402 cells, which also indicated that Stealth RNAi against GP73 (RNAi) mainly caused accumulation of early-stage apoptotic Bel7402 cells, however, many vacuoles, nuclear fragmentation and apoptotic bodies appeared in Hep G2 cells, which can also explain why there was an apoptotic peak in the RNAi group. These results also confirmed that Stealth RNAi against GP73 (RNAi) mainly caused accumulation of terminal-stage apoptotic Hep G2 cells. And the difference of its expression in different cells might play different function. Several studies have found that the ratio of Bax/Bcl-2 is important in apoptosis of cancer cells. Bcl-2 is considered as an upstream effector molecule in the apoptotic pathway, but Bax is considered as a downstream effector molecule[36,37]. The protein levels of Bax and Bcl-2 were measured, and similar results were also found in this study. We demonstrated that the reduction of GP73 led to a decrease in expression of Bcl-2 but an increase in Bax, and the Bcl-2/Bax ratio in Hep G2 cells was reduced, this result was also similar to the other one on the cell apoptotic mechanism[38]. Besides, in some apoptotic mechanism studies, it was found that Bcl-2 inhibits cytochrome c release from mitochondria, whereas Bax triggers it[39], and release of cytochrome c activated caspases to induce apoptosis. The expression of cytochrome c and procaspase-3 was also detected by Western blotting to determine their distribution in apoptosis. The level of cytochrome c in the cytoplasm was markedly elevated in the RNAi group; however, the level of procaspase-3 was reduced significantly. These results proved that GP73 may play a major role in anti-apoptosis in HCC cells, and there is a correlation between it and other apoptosis-related proteins such as Bax, Bcl-2, cytochrome c and procaspase-3.
In conclusion, Stealth RNAi targeting GP73 gene sequence reduced the expression of GP73 markedly. The reduction of GP73 in Hep G2 and Bel7402 cells inhibited cell proliferation and induced apoptosis, however, terminal apoptosis occurred in Hep G2 cells, but early apoptosis occurred in Bel7402 cells. Reduced expression of GP73 gene might lead to a reduction in Bcl-2/Bax ratio, an increase in cytochrome c, but a reduction in capase-3, and the reduction of GP73 induces apoptosis in Hep G2 cells may through this apoptosis signaling pathway. However, further studies are needed to confirm this conclusion. Consequently, the results imply that GP73 plays an important role in HCC cells, but unfortunately, expression of GP73 was not reduced to a low level in this study, therefore, more effective molecular tools are needed to explore the exact function of GP73 in more HCC cells.
COMMENTS
Background
The physiological and pathological roles of Golgi protein (GP)73 have attracted considerable attention in recent years. However, the function of GP73 in hepatic carcinoma cells remains obscure.
Research frontiers
This study was performed to explore the function of GP73 in hepatic carcinoma cells; Stealth RNAi targeting GP73 gene sequence was used to silence its expression; and cell proliferation and apoptosis were assessed. This study implied that GP73 plays an important role in hepatocellular carcinoma (HCC) cells.
Innovations and breakthroughs
The results of this study suggested that silencing GP73 gene in HCC cells inhibits cell proliferation and induces apoptosis.
Applications
The results imply that GP73 plays an important role in HCC cells, but further research is needed to explore the exact function of GP73 in more HCC cells.
Peer review
These results are very interesting and may provide important evidence about the mechanism of apoptosis in HCC.