Copyright
©2014 Baishideng Publishing Group Inc.
World J Gastroenterol. Aug 7, 2014; 20(29): 9912-9921
Published online Aug 7, 2014. doi: 10.3748/wjg.v20.i29.9912
Published online Aug 7, 2014. doi: 10.3748/wjg.v20.i29.9912
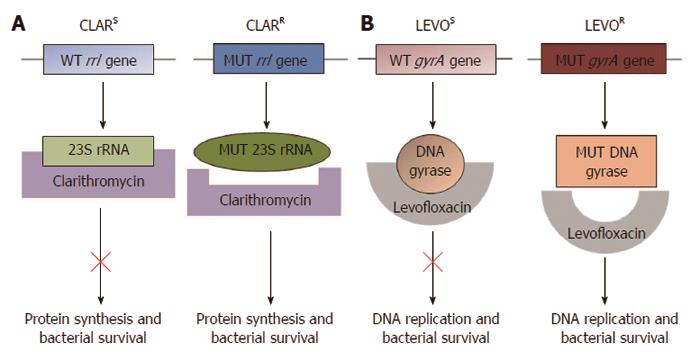
Figure 1 Molecular mechanism of clarithromycin and levofloxacin resistance.
A: Clarithromycin binds and suppresses the activity of the bacterial ribosomal subunit, thus inhibiting protein synthesis. Single point mutations within the Helicobacter pylori (H. pylori) rrl gene encoding the 23S ribosomal RNA component of ribosomes result in clarithromycin resistance. The best characterised point mutations conferring clarithromycin resistance are A2146C, A2146G and A2147G; B: Levofloxacin targets H. pylori DNA gyrase. The most significant mutations conferring levofloxacin resistance occur at positions 87 and 91 of the H. pylori gyrA gene, which encodes the A subunit of the DNA gyrase enzyme involved in regulating the topological state of bacterial DNA during replication. CLARS: Clarithromycin sensitive; CLARR: Clarithromycin resistant; LEVOS: Levofloxacin sensitive; LEVOR: Levofloxacin resistant; WT: Wild type; MUT: Mutant.
-
Citation: Smith SM, O’Morain C, McNamara D. Antimicrobial susceptibility testing for
Helicobacter pylori in times of increasing antibiotic resistance. World J Gastroenterol 2014; 20(29): 9912-9921 - URL: https://www.wjgnet.com/1007-9327/full/v20/i29/9912.htm
- DOI: https://dx.doi.org/10.3748/wjg.v20.i29.9912