Copyright
©2014 Baishideng Publishing Group Inc.
World J Gastroenterol. Jul 21, 2014; 20(27): 8928-8938
Published online Jul 21, 2014. doi: 10.3748/wjg.v20.i27.8928
Published online Jul 21, 2014. doi: 10.3748/wjg.v20.i27.8928
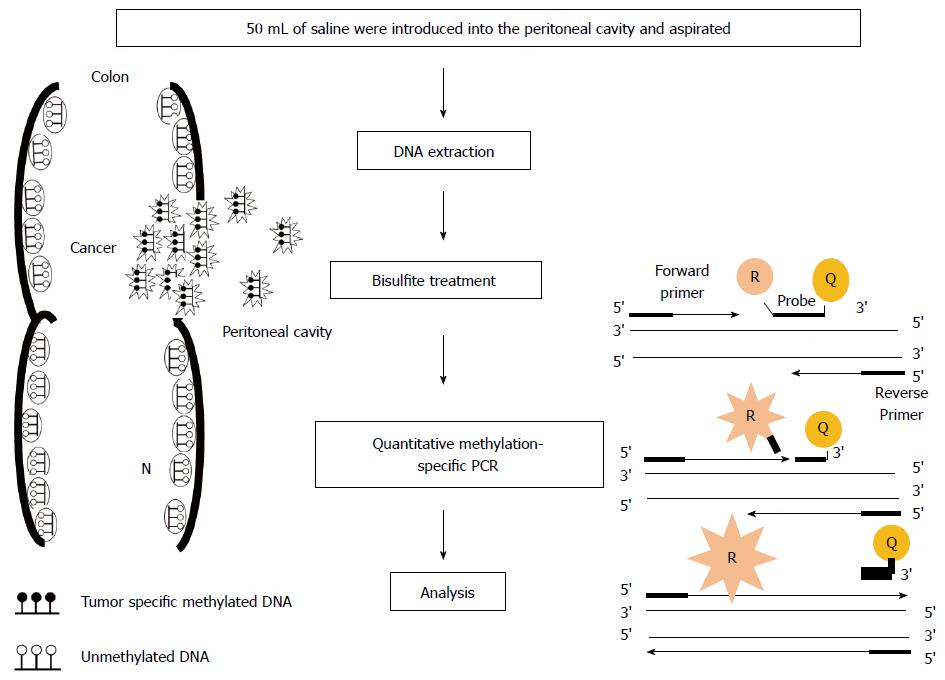
Figure 1 Outline of quantitative methylation-specific polymerase chain reaction analysis in peritoneal lavage.
The depth of invasion and subsequent increased likelihood of tumor cells exfoliated from colonic serosa is reflected in the presence of tumor-related methylated DNA in peritoneal cavity. In quantitative methylation-specific polymerase chain reaction (PCR) step, the DNA polymerase cleaves only probes that are hybridized to the target. Cleavage separates the reporter dye (R) from the quencher dye (Q); resulting in increased fluorescence by the reporter. The increase in fluorescence signal occurs only if the target sequence is complementary to the probe and is amplified during PCR.
- Citation: Kamiyama H, Noda H, Konishi F, Rikiyama T. Molecular biomarkers for the detection of metastatic colorectal cancer cells. World J Gastroenterol 2014; 20(27): 8928-8938
- URL: https://www.wjgnet.com/1007-9327/full/v20/i27/8928.htm
- DOI: https://dx.doi.org/10.3748/wjg.v20.i27.8928