Copyright
©2014 Baishideng Publishing Group Inc.
World J Gastroenterol. Jun 21, 2014; 20(23): 7137-7151
Published online Jun 21, 2014. doi: 10.3748/wjg.v20.i23.7137
Published online Jun 21, 2014. doi: 10.3748/wjg.v20.i23.7137
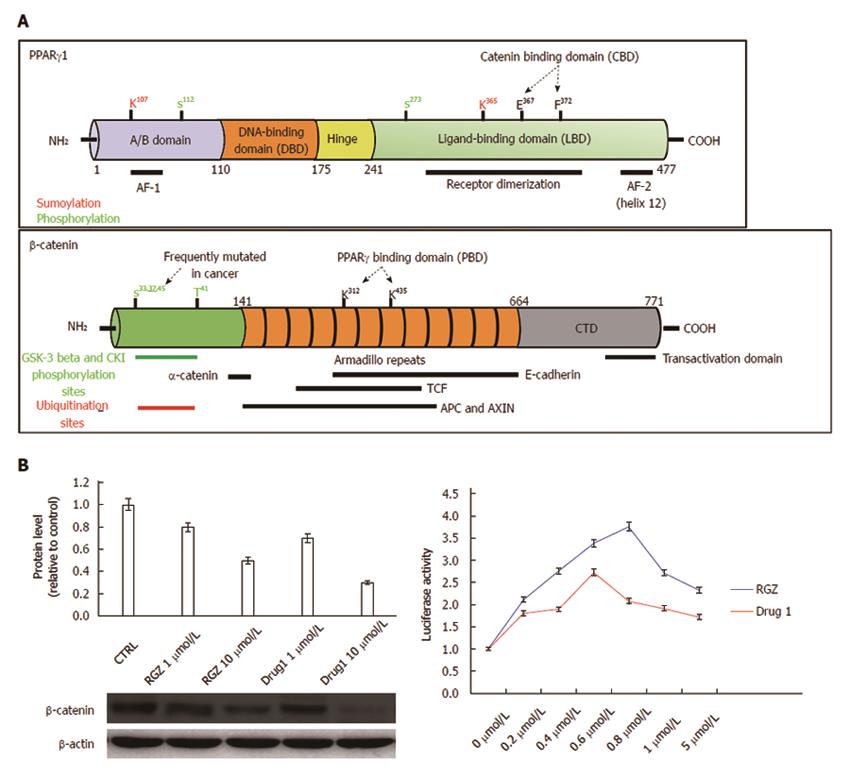
Figure 3 Structural and functional domains of PPARγ and β-catenin.
A: The mature PPARγ protein consists of four structural/functional domains: (1) the variable A/B region at the N terminus contains the ligand-independent transactivation domain AF1 (residues 1–71 of PPARγ1); lysine 79 and serine 84 residues are targets of SUMOylation and phosphorylation events, respectively; (2) the C region is the DNA binding domain, characterized by two C4 Zinc-finger motifs, that interact with the major groove of the DNA; (3) the D or hinge region allows receptor dimerization and DNA binding; and (4) the E/F region is the ligand binding domain (LBD) constituted by 12 α-helices and 4 β-strands where the agonist accommodates. This region (helices 7 and 8) includes a β-catenin binding domain (CBD) essential for the interaction with β-catenin. The most important aminoacid residues implicated in PPARγ activity regulation are shown. The full length β-catenin is essentially composed by three domains: (1) the N-terminal domain involved in the ubiquitin-mediated degradation; (2) the arm repeat domain, containing 12 armadillo repeats that mediate the binding with cadherins, APC, TCF/LEF, CREB binding protein (CBP) and PPARγ; and (3) the carboxy terminal (CTD) or transactivating domain interacts with coactivators such as CBP or corepressors such as β-catenin inhibitor and TCF-4 (ICAT). The most important aminoacid residues implicated in β-catenin activity regulation are shown; B: Luciferase activity from HEK293T cells transfected with a PPRE-driven luciferase reporter gene and exposed to the compound indicated as Drug1 is lower than that obtained from cells exposed to rosiglitazone, indicating a reduced transactivation potential in line with the notion of a partial agonist. HT29 colon cancer cells treated with Drug1 exhibit inhibition of cell growth and a 40% higher ability to downregulate β-catenin than rosiglitazone, likely through a mechanism involving β-catenin nuclear export and proteasome-mediated degradation.
- Citation: Sabatino L, Pancione M, Votino C, Colangelo T, Lupo A, Novellino E, Lavecchia A, Colantuoni V. Emerging role of the β-catenin-PPARγ axis in the pathogenesis of colorectal cancer. World J Gastroenterol 2014; 20(23): 7137-7151
- URL: https://www.wjgnet.com/1007-9327/full/v20/i23/7137.htm
- DOI: https://dx.doi.org/10.3748/wjg.v20.i23.7137