Published online Jun 21, 2014. doi: 10.3748/wjg.v20.i23.7104
Revised: December 5, 2013
Accepted: April 1, 2014
Published online: June 21, 2014
Hepatitis C virus (HCV) is a major cause of chronic liver diseases, including steatosis, cirrhosis and hepatocellular carcinoma, and its infection is also associated with insulin resistance and type 2 diabetes mellitus. HCV, belonging to the Flaviviridae family, is a small enveloped virus whose positive-stranded RNA genome encoding a polyprotein. The HCV core protein is cleaved first at residue 191 by the host signal peptidase and further cleaved by the host signal peptide peptidase at about residue 177 to generate the mature core protein (a.a. 1-177) and the cleaved peptide (a.a. 178-191). Core protein could induce insulin resistance, steatosis and even hepatocellular carcinoma through various mechanisms. The peptide (a.a. 178-191) may play a role in the immune response. The polymorphism of this peptide is associated with the cellular lipid drop accumulation, contributing to steatosis development. In addition to the conventional open reading frame (ORF), in the +1 frame, an ORF overlaps with the core protein-coding sequence and encodes the alternative reading frame proteins (ARFP or core+1). ARFP/core+1/F protein could enhance hepatocyte growth and may regulate iron metabolism. In this review, we briefly summarized the current knowledge regarding the production of different core gene products and their roles in viral pathogenesis.
Core tip: In addition to the mature core protein (a.a. 1-177) and the cleaved peptide (a.a. 178-191), different alternative reading frame (ARF)/core+1 proteins could be expressed from the core+1 reading frame of hepatitis C virus (HCV) core gene. Core gene products play an important role in the HCV pathogenesis. Core protein could induce insulin resistance, steatosis, and even hepatocellular carcinoma. The peptide (a.a. 178-191) may play a role in the immune response and steatosis development. ARF proteins/core+1/F protein could enhance hepatocyte growth and may regulate iron metabolism. We summarized the current knowledge regarding the HCV core gene products and their pathogenicity in this article.
- Citation: Li HC, Ma HC, Yang CH, Lo SY. Production and pathogenicity of hepatitis C virus core gene products. World J Gastroenterol 2014; 20(23): 7104-7122
- URL: https://www.wjgnet.com/1007-9327/full/v20/i23/7104.htm
- DOI: https://dx.doi.org/10.3748/wjg.v20.i23.7104
Hepatitis C virus (HCV) accounts for approximately 15%-20% cases of acute hepatitis. After acute infection, around 50% to 80% of HCV patients will develop chronic infection. HCV persistently infected individuals are at risk to develop liver inflammation, steatosis, fibrosis, cirrhosis and hepatocellular carcinoma (HCC)[1-4]. Epidemiological studies also indicate that HCV is associated with insulin resistance and type 2 diabetes mellitus[5,6].
HCV is a small enveloped RNA virus belonging to the family Flaviviridae and genus hepacivirus. The HCV genome is a single, positive-stranded RNA with a nucleotide length of about 9.6 kb. This genome encodes a polyprotein precursor of approximately 3000 amino acids, which is processed by host and viral proteases into at least 10 different proteins, which are arranged in the order of NH2-C-E1-E2-p7-NS2-NS3-NS4A-NS4B-NS5A-NS5B-COOH. C, E1, and E2 are structural proteins while NS2-NS5B and perhaps also p7 are non-structural proteins. The release of C, E1, E2 and, p7 from the polyprotein is mediated by the cellular signal peptidase located in the endoplasmic reticulum, whereas the cleavages between NS2-NS5B are mediated by viral NS2/3 and NS3/4A proteases (for a review[7,8]).
Following the discovery of HCV, the presence of great nucleotide diversity among isolates was reported[1,9]. Due to the lack of proof-reading mechanism in the NS5B polymerase, a closely related but diverse population of viral variants known as quasispecies is produced at a rate of approximately one mutation per replication cycle within infected individuals[10]. Accumulation of nucleotide substitutions in the virus has resulted in diversification into subtypes and distinct genotypes. Therefore, the HCV RNA genome sequences are highly heterogeneous. At present, HCV is classified into seven major genotypes and numerous subtypes[11]. There is 30%-50% variation among viral genotypes and 15%-30% variation among different subtypes, while there is 1%-5% variation in HCV nucleotide sequence from a single infected patient[12,13]. Viral pathogenesis and response to anti-viral treatment are different among different HCV genotypes, e.g., genotype 3 infection is associated with a high level of liver steatosis while genotypes 1 and 4 are more resistant to interferon (IFN) based therapies than genotypes 2 and 3[11].
The pathobiological changes caused by HCV infection have been attributed to both the host immune responses and the direct viral cytopathic effects[8]. Viral pathogenesis caused by direct viral cytopathic effects is the outcome of the interactions between the host cell and different HCV proteins. In this review, we will only focus on the pathogenicity of HCV core gene products.
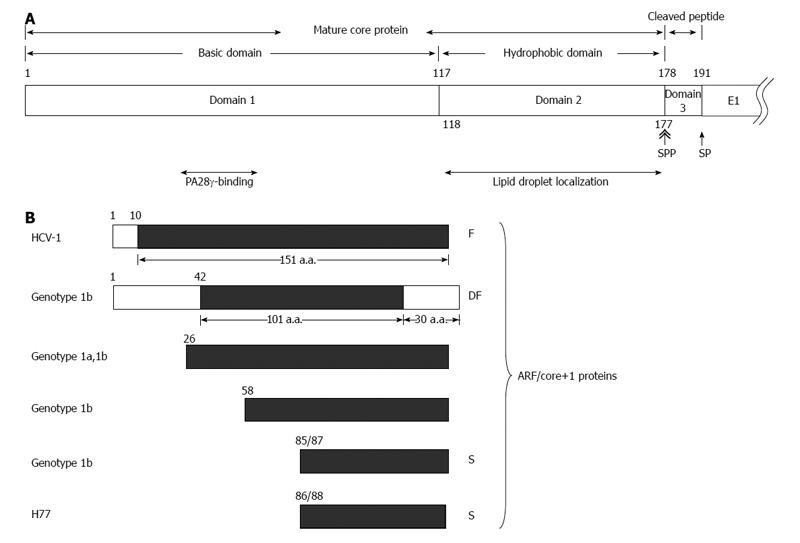
The HCV core protein is cleaved at residue 191 by the host signal peptidase (SP) to release it from the precursor polyprotein. This immature core protein is further cleaved by the host SP peptidase (SPP) within the C-terminal transmembrane region to generate the mature core protein and the cleaved peptide. The cleavage of HCV core protein by SPP is essential for HCV assembly[14,15]. The cleavage between core and E1 proteins by SP facilitates further cleavage of core protein by SPP[16]. The exact C terminal amino acid residue of the mature, virion-associated core protein is not known. The studies of core protein expressed in insect cells have indicated that SPP cleaves after residue 177[17], 179 or 182[18], and the studies in human cells have supported that the mature protein terminates at residue 177[15]. Moreover, a recent report demonstrated that a core protein with amino acids 1 to 177 efficiently trans-complemented the viral assembly[19]. Therefore, although the exact C terminus of the core protein awaits further investigation, it is likely that the mature core protein contains 177 amino acid residues. And, the cleaved peptide generated by the sequential cleavages of SP and SPP is composed of amino acid 178 to 191 (Figure 1A).
Comparing with other HCV proteins, core protein is thought to be the most conserved one: results of nucleotide and deduced amino acid sequence analysis across diverse HCV strains reveal 81%-88% nucleotide and 96% amino acid sequence homology[20,21]. Assembly of the virion is initiated by the oligomerization of core protein. Small molecules directly binding to core protein could potentially be potent antiviral agents[22].
The mature core protein having 177 amino acids consists of two domains: positively charged domain 1 (a.a. 1-117) and hydrophobic domain 2 (a.a. 118-177) (Figure 1A). The domain 1 is rich in basic residues, and is implicated in RNA-binding and homo-oligomerization. The domain 2 is important for the membrane association activity of the core protein as well as for its folding and stability (for a review[23,24]). The amphipathic helices I and II in domain 2 spanning from residue 119 to 136 and residue 148 to 164, respectively, are involved in the association of HCV core protein with lipid droplets[25]. In addition, the region spanning from residue 112 to 152 is associated with membranes of the endoplasmic reticulum and mitochondria[26,27].
The core protein may also localize with nucleus[26,28] and bind to the nuclear proteasome activator PA28-γ/REGγ, resulting in PA28-γ-dependent degradation of the core protein[28].
The cleaved peptide (a.a. 178-191) is highly conserved with close to 100% identity among different HCV genotypes[19,29].
In addition to the proteins translated from the conventional open reading frame (ORF), existence of a new antigen encoded in the -2/+1 alternative reading frame (named ARF) was first demonstrated through bioinformatics and patient-based research in 2001[30].
Besides the core protein, a smaller protein was first detected in 1994 when the HCV-1 isolate was in vitro translated[31]. In 2001, this smaller protein (named F) was found to be synthesized by the ribosomal frameshift into the -2/+1 reading frame, not from the conventional viral open reading frame. Results from sequence analysis of different HCV genotypes and from the reactivity of patients’ sera also indicated the existence of a protein product encoded from this -2/+1 reading frame[32].
In 2002, translation of this -2/+1 reading frame (named core+1) was also verified by the fusion protein reporter assay and antibody response[33]. Therefore, proteins from this alternative reading frame was named ARFP/F/core+1 at first (for a review[34]).
In addition to F protein, proteins with different lengths were synthesized from this alternative reading frame through various translational mechanisms used in different HCV genotypes and/or strains. The ARFP/double-frameshift (DF) protein from genotype 1b is a chimera: N-terminal with 42 amino acids of the core protein, followed by 101 amino acids of ARFP in the middle, and end with the C-terminal 30 amino acids of the core protein[35]. Translation from the ARF could be started from non-AUG codon 26 (core+1), GUG or GCG, of genotype 1a or 1b, or initiated from codon 58 (GUG) of genotype 1b[36,37]. Actually, translation initiation from this ARF was detected most efficient at the internal AUG codon at position 85/87 of genotype 1b or 86/88 of H77 strain[37-39], named core+1/S (short form). Therefore, proteins translated from the +1 reading frame are composed of proteins with different lengths[34,40] (Figure 1B). All proteins containing amino acids from this ARF are called ARFP, core+1 proteins or ARF/core+1 proteins. Specific proteins from this ARF are designated after ARFPs or core+1, e.g., F protein was called ARFP/F, core+1/F or ARFP/core+1/F[34,41].
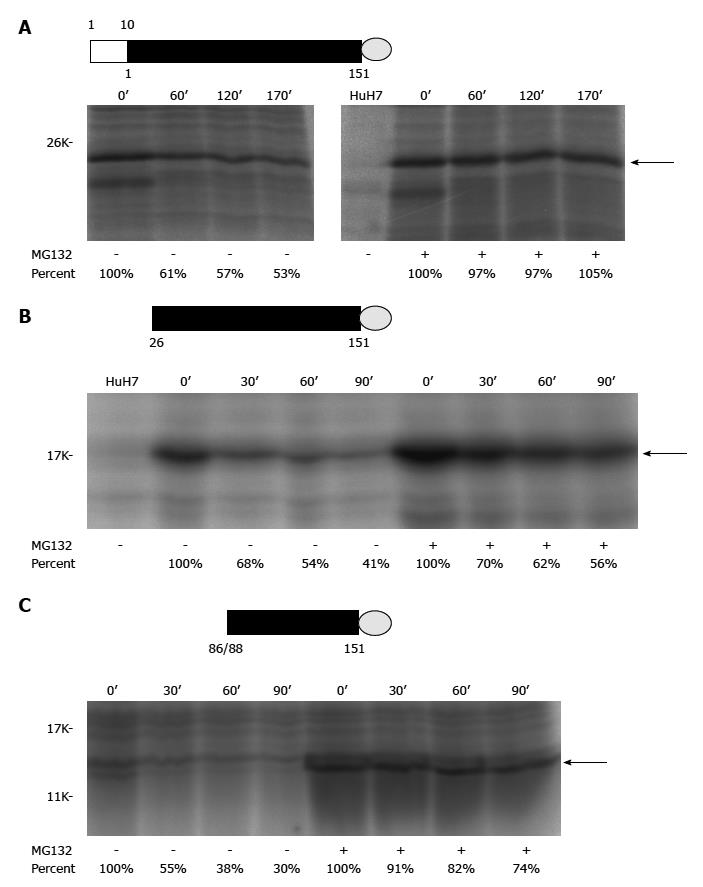
Different ARF/core+1 proteins could have similar subcellular localization, e.g., both F and core+1/S are cytoplasmic proteins, primarily associated with the endoplasmic reticulum[37,42]. Further immunoflurescence and subcellular fractionation analyses indicated that core+1/S and core+1/F are cytoplasmic proteins with partial endoplasmic reticulum distribution at interphase, whereas in dividing cells they also localize to the microtubules of the mitotic spindle[41]. ARF/core+1 proteins seem to be labile in the cells. F protein was labile in the cells and its degradation is ubiquitin-independent[42,43]. Moreover, core+1/S is also very unstable[41]. In the cells, the half-lives of several ARF/core+1 proteins were around 30 to 120 min (Figure 2A-C). Biochemical properties of different ARF/core+1 proteins are largely unknown. Core+1/S, a highly basic polypeptide, was found to be highly disordered under native conditions, with a tendency for self-association[44].
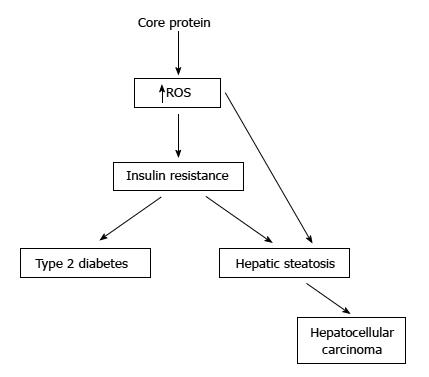
At present, the transgenic mouse model was used mostly to study the pathogenic roles of core protein in animals[45]. Core protein was shown to induce the ROS overproduction in the liver of transgenic mice[46]. In one study, core protein could induce steatosis only in the transgenic mice[47]. In another study, the transgenic mice expressing core protein developed steatosis and HCC in the absence of inflammation[48]. On the other hand, the transgenic mice with constitutive core protein expression developed insulin resistance at 1 to 2 mo-old, then leading to type 2 diabetes on a high-fat diet. Most of these mice would develop hepatic steatosis at 6-mo-old and some of them would develop HCC at 16 to 23 mo-old[24,49-51] (Figure 3).
Transgenic mice with constitutive expression of core protein are usually lack of immune response to this protein. Therefore, transgenic mouse models suitable to study fibrosis and cirrhosis caused by core protein are not available yet. A Cre/loxP recombination system has been developed in transgenic mice to study the inflammation caused by the core protein[52]. This inducible system in transgenic mice may be suitable to study fibrosis and cirrhosis caused by core protein in the near future.
Molecular mechanisms regarding the pathogenic roles of core protein were studied extensively in the cell culture and transgenic mouse models.
HCV proteins orchestrate a complex and dynamic interaction network with cellular proteins contributing to viral persistence and pathogenecity. Through high-throughput yeast two-hybrid screening assay and computation-based analysis, a virus-human protein interactome network has been constructed[53]. Cellular proteins related to four pathways are major targets by HCV proteins: insulin, Jak/STAT, TGF-β and focal adhesion pathways. Core protein appeared as a major perturbator of IJT network (insulin, Jak/STAT and TGF-β pathways) in this study. Seventy-six cellular proteins were found to interact with core protein in this yeast two-hybrid screening assay. By interacting with PLSCR1, connecting insulin and JAK/STAT, core protein could therefore interfere with both insulin and JAK/STAT pathways. Through interacting with Yin Yang 1, connecting IJT network, core protein could perturb these three pathways[53]. To explore the protein-protein interactions further, the yeast two-hybrid membrane protein system was performed. Eleven human proteins interacting with core protein were identified in this assay. A virus-human protein interactome network has also been constructed[54]. This network suggests that core protein may (1) interfere the host innate immune response through interacting with SLC25A5; (2) induce oxidative stress through interacting with NDUFS2 and ETFB; (3) affect focal adhesion pathway through SLC25A5 and ENO1; and (4) elevate hepatic iron level through its cellular partner FTL[54]. Therefore, core protein could potentially target insulin, Jak/STAT, TGF-β and focal adhesion pathways.
Through extensive literature review, more than 100 cellular proteins (including the proteins mentioned above) were found to interact with core protein and the interaction network of core protein was constructed. These cellular proteins are involved in the processes of signal transduction, transcription, nucleic acid binding, apoptosis, cell cycle, cytoskeleton and kinase activity[55]. Pathogenicity of core protein may be resulted from its interaction with these cellular proteins.
HCV core protein could modulate cellular gene expression by directly interacting with transcription factors or indirectly through affecting the signal transduction pathways. Expression of numerous cellular genes is regulated by HCV core protein (for a review[56]). This review focuses only on the cellular genes modulated by core protein identified through global analysis, i.e., microarray. Core protein could stimulate hepatocyte growth, which is at least partly mediated through up-regulation of Wnt-1 expression, both in Huh-7 cells and transgenic mice[57,58]. Stat3 signaling pathway was induced when primary human hepatocytes was immortalized by core protein[59]. Genes involved in lipid metabolism (e.g., SREBP pathway) were affected by core protein in either cultured cells or transgenic mice[60,61]. Core protein could also induce interferon-inducible gene 27 in primary human hepatocytes[62]. Moreover, transgenic mice that conditionally express intermediate HCV core protein develop inflammation possibly through activation of complement 3[52]. On the other hand, core protein may mute the cellular inflammatory response via inhibition of cyclooxygenase 2 expression during HCV infection[63]. In B cells, core protein may impair antigen presentation by downregulation of MHC class II molecules[64]. Therefore, gene expression profiles regulated by core protein are mainly involved in lipid metabolism, signal transduction, protease activity and immune responses[59,60,65-67]. It is important to notice that cellular gene expression profiles modulated by core proteins from different genotypes are not the same[60,67].
MicroRNAs (miRNAs) affect gene silencing via translational inhibition and/or mRNA degradation[68]. The miRNA dysfunction is believed to play important roles in human diseases, including viral infectious diseases, e.g., HCV infection (for a review[56]). HCV core protein could down-regulate the expression of miRNA-122 and miRNA-124 in the cells[69,70]. On the other hand, core protein could down-regulate p21(Waf1/Cip1) expression by enhancing miRNA-345 expression in human hepatoma cells[71]. In monocytes, core protein could also increase the miRNA-155 expression, which in turn up-regulate the TNF-α production[72]. Recently, core protein was reported to induce steatosis through up-regulation of the miRNA-27 expression[73]. It is not surprising to know that differentially expressed microRNAs were detected in HuH-7 cells expressing core proteins from different genotypes[74].
Pathogenicity of core protein should be at least partially through modulating cellular gene expression (mRNA and/or miRNA production).
Apoptosis is important for a host to defend viral infections, to inhibit viral spread and persistence. Induction of apoptotic pathways in HCV-infected patients, primarily as a result of host immune responses, could lead to viral suppression and virus-mediated liver damage. In HCV-infected liver, however, despite enhanced hepatocyte apoptosis, viral persistence is observed. To date, it is not known whether the infectious virions act as pro- or anti-apoptotic agent in vivo. For virtually all HCV proteins, pro- and anti-apoptotic effects have been described. However, which HCV protein affecting apoptosis in vivo is still unknown (for a review[56,75,76]).
The data regarding the effect of HCV core protein on apoptosis is controversial. Core protein was reported to enhance Fas-mediated apoptosis[77]. However, Fas-mediated apoptosis was inhibited in the transgenic mice expressing core, E1, E2 and NS2 proteins[78]. Core protein could either enhance or inhibit TNFα-mediated apoptosis[79-81]. The discrepancy of these results is possibly due to that different virus strains and/or different experimental systems were used. Recently, core protein from genotype 3a was demonstrated to have a stronger effect on anti-apoptosis than the one from genotype 1a[82]. Moreover, it has been mentioned that other factors, possibly cell-type specific, might be involved in different pro- and anti-apoptotic effects of core protein in the cells[83]. Therefore, it is unclear whether core protein inhibits or induces apoptosis in hepatocytes. However, it is still believed that inhibition of apoptosis and enhanced cell proliferation are important in progression of HCC[76]. Recently, core protein was reported to have anti-apoptotic effect in B cells that were isolated from two individual donors[64].
Through escaping immune detection and/or suppressing the host immune responses, HCV is efficient to establish persistent infection[83]. HCV core protein could modulate immune response in many ways. It is well known that core protein would suppress interferon signaling (for a review[84]). Core protein is reported to block interferon signaling by interacting with the STAT1 protein[85,86]. Through inhibition of interferon regulatory factor-3 (IRF-3) dimerization, core protein suppressed interferon β expression[87]. By reducing the interaction with DEAD-box RNA helicase (DDX3), core protein with specific mutations also attenuated type I interferon response[88]. Core protein is also reported to abrogate DDX3 and interfere with DDX3-enhanced interferon signaling in two different cells[89]. In addition, core protein may alter NK cell function by inducing apoptosis in these cells[90]. Core protein was also reported to stimulate TLR2 pathway assisting the virus to evade from the innate immune system[91]. Therefore, core protein impairs the innate immunity through these activities.
In hepatocytes, HCV infection or core protein expression could up-regulate the CD55 expression, limiting excessive complement activation[92]. Moreover, hepatocytes infected with HCV or expressing core protein displayed significant repression of complement 9 expression[93]. On the contrary, in the transgenic mice with core protein, complement 3 was up-regulated in inflamed liver. Moreover, administration of CD55 reduced hepatic inflammation[52]. It is not known which factor(s) caused the discrepancy. However, it is possible that core protein may modulate the complement activity in different situations.
In the transgenic mice, core protein could suppress T cell response via enhancing Fas-mediated apoptosis in these cells[94]. In addition, through interaction with gClqR, core protein could up-regulate the expression of PD-1 and SOCS-1 in T cells and monocytes/macrophages[95,96], of Tim-3 in monocytes[97], and of STAT3 on human monocytes, macrophages, and dendritic cells[98], and in turn, suppress the functions of these cells. Core protein could also inhibit cathepsin S-mediated MHC class II maturation, contributing to weak immunogenicity of viral antigens in chronically infected humans[99]. Therefore, core protein affected the innate and adaptive immunities.
Jurkat cells expressing core proteins suppressed CD4+ and CD8+ T-cell responses to anti-CD3 plus anti-CD28 stimulation by up-regulation of both FOXP3 and CTLA-4 expression[100]. Therefore, core protein also inhibited the functions of regulatory T cells.
In summary, HCV core protein could modulate immune responses through different mechanisms.
As mentioned earlier, core protein is known to inhibit interferon signaling[84-89]. Therefore, core protein sequence variation may be associated with interferon (IFN) therapy resistance. Indeed, substitutions of amino acid 70 and amino acid 91 in the core protein of genotype 1b were reported as independent factors associated with a non-virological response toward interferon treatment. Especially, substitutions of arginine by glutamine at amino acid 70, and/or leucine by methionine at amino acid 91 were significantly more common in non-virological responses toward interferon treatment[101]. After this finding, numerous reports confirmed this association (for a review[11,84]). Consistent with these clinical findings, an in vitro study has also demonstrated that cells with core mutants (R70Q, R70H, and L90M) were significantly more resistant to the interferon treatment than the cells with the wild-type core protein. Moreover, the interferon-resistance of the cells with these core mutants may be through IL-6-induced, SOCS3-mediated suppression of interferon signaling[102].
HCV infection is characterized by a systemic oxidative stress. The possible mechanisms of HCV-induced oxidative stress include (1) activation of NAD(P)H oxidase of Kupffer cells and PMN cells during inflammation; (2) iron overload and lipid peroxidation; (3) activation of NAD(P)H oxidase by NS3 protein; (4) increased production of mitochondrial ROS/RNS by the electron transport chain due to core and NS5A proteins; (5) decreased GSH output due to liver damage; (6) decreased antioxidants and antioxidant gene expression; (7) alcohol, drugs, and other chemicals; (8) increased cytokines that increase ROS; (9) increased expression/activity of COX-2; and (10) increased expression of CYP2E1 (for a review[103]). HCV core protein is reported to be associated with endoplasmic reticulum (ER) and interacted directly with mitochondria. Then, core protein would cause ER stress, inhibit mitochondrial electron transport and increase ROS production[104,105].
In the transgenic mice, core protein could induce overproduction of ROS in liver. At the same time, some genes of the antioxidant systems, including heme oxygenase-1 and NADH dehydrogenase quinone 1, were down-regulated in the liver with HCV infection[46]. Similarly, in cooperation with NS3 protein, core protein would impair the induction of cytoprotective Nrf2 target genes in the cells[106]. The expression of a variety of cytoprotective genes is regulated by short cis-acting elements in their promoters, called antioxidant response elements (AREs). A central regulator of ARE-mediated gene expression is Nrf2. Therefore, core protein could induce ROS production and impair cytoprotective response.
On the other hand, it was reported that core protein expression leads to intracellular oxidative stress, and that vital cellular functions are, in turn, protected by the up-regulation of cellular antioxidant defense mechanisms in cultured hepatoma cells[107]. Furthermore, core protein could activate the antioxidant defense Nrf2/ARE pathway in a ROS-independent manner[108]. Therefore, core protein could induce ROS production and, at the same time, activate the cytoprotective response.
In summary, core protein could induce ROS production. Meanwhile, core protein might up- or down-regulate Nrf2 target genes in different conditions.
HCV infection is known to be associated with insulin resistance (IR)[109], leading to the development of type 2 diabetes[6,110]. IR in chronic hepatitis C is reported to associate with genotypes 1 and 4, the serum HCV RNA level, and liver fibrosis[111]. Therefore, IR could be promoted by HCV via a genotype-specific mechanism. HCV core protein is a pathogenic factor for the development of IR[84,112,113].
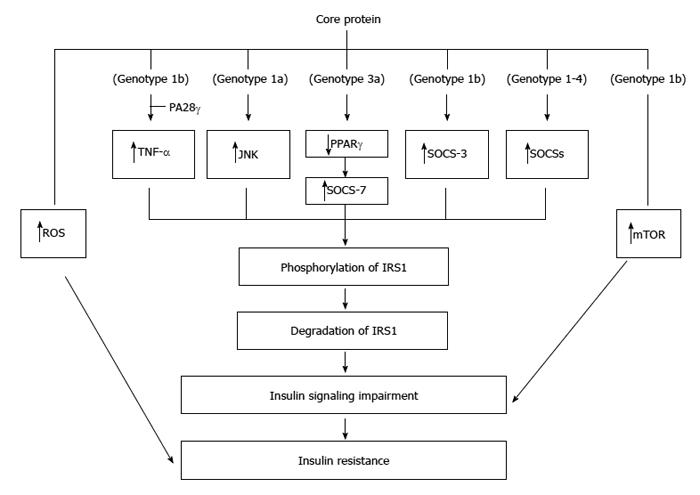
Core protein could cause ER stress, increase oxidative stresses[104,105], which can further exacerbate IR[114]. IR caused by core protein (genotype 1b) in transgenic mice was at least partially mediated by induction of TNF-α over-production, responsible for phosphorylation of serine residues of insulin receptor substrates (IRS-1 and IRS-2) and down-regulation of glucose transporter gene expression. Indeed, administration of antibodies against TNF-α to these mice could restore insulin levels to normal and return insulin sensitivity to normal[51]. Further analysis of this mouse model indicated that a PA28-γ-dependent pathway was required for core protein-mediated IR[49]. To impair the insulin signaling, core protein (genotype 1a) increases IRS-1 phosphorylation at Ser(312) by activating JNK in hepatocytes[115] (Figure 4).
Core proteins from different genotypes down-regulate IRS-1 through genotype-specific mechanisms: the core protein of genotype 3a promoted IRS-1 degradation through the down-regulation of PPAR-γ and by upregulating SOCS-7 while the core protein of genotype 1b activated mTOR[116]. Further study indicated that core protein of genotype 3a increases SOCS-7 expression through PPAR-γ in Huh-7 cells[117]. Indeed, deletion of SOCS-7 in the transgenic mice leads to enhanced insulin action[118]. In addition to SOCS-7, over-expression of SOCS-3 has also been linked to insulin resistance[119]. Indeed, core protein (genotype 1b) up-regulated SOCS-3 and caused ubiquitination of IRS1 and IRS2 in the transgenic mice. Furthermore, core protein-induced down-regulation of IRS1 and IRS2 was not seen in the SOCS3(-/-) mouse embryonic fibroblast cells[120]. Actually, activation of SOCS family members is a general mechanism associated with the core proteins from genotypes 1-4 except a rare genotype 1b variant failed to activate any of the SOCS tested. This leads to identifying the role of the amino acids 49 and 131 of core protein in mediating SOCS transactivation[121].
Sequence variations in core protein may affect IR development. Indeed, in Japanese patients without cirrhosis and diabetes mellitus, a.a. substitutions of the genotype 1b core protein [Glu70 (His70) and/or Met91] are the significant determinants of severe IR[122].
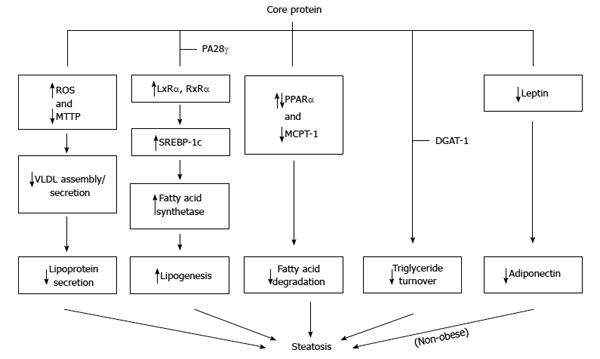
Steatosis or ‘‘fatty liver’’ is common in patients infected with HCV. Steatosis caused by HCV infection should be classified into two types: metabolic steatosis in patients infected with non-genotype 3, and viral steatosis in patients infected with genotype 3. Metabolic steatosis occurs in the setting of obesity, hyperlipidemia, and IR, whereas viral steatosis is caused by HCV as a direct cytopathic effect[112,123-126]. Transgenic mice with core protein developed hepatic steatosis[47,127]. Therefore, core protein plays an important role in the development of steatosis[128]. Three mechanisms have been proposed regarding the induction of triglyceride accumulation in the liver cells by core protein: firstly by impaired lipoprotein secretion, secondly by increased lipogenesis, and thirdly by impaired fatty acid degradation[124-126,128] (Figure 5).
To impair lipoprotein secretion, core protein inhibits the activity of microsomal triglyceride transfer protein (MTTP), which plays a key, rate-limiting role in VLDL assembly/secretion. Thus, its inhibition results in the accumulation of triglycerides in the cells, which causes steatosis[129]. Furthermore, through interaction with mitochondria, core protein induces ROS production. The production of ROS results in peroxidation of membrane lipids and structural proteins, that are involved in the trafficking and secretion apparatus, blocking VLDL secretion.
To increase lipogenesis, core protein activates transcription factor SREBP-1c through up-regulation of liver X receptor alpha (LXRα) and retinoid X receptor alpha (RXRα), leading to enhanced activity of various enzymes involved in cellular lipid biosynthesis[61,130-133]. Interestingly, the genes related to fatty acid biosynthesis and srebp-1c promoter activity were up-regulated by core protein in cell line and mouse liver in a PA28γ-dependent manner[50]. Recently, a study reported that accumulation of triglycerides in HepG2 cells with core proteins was due to delta-9 desaturase, an enzyme involved in fatty acid biosynthesis (primarily the synthesis of oleic acid), activated by core protein. Moreover, polyunsaturated fatty acids could counteract the impact of core protein on lipid metabolism[134].
To impair fatty acid degradation, core protein is believed to down-regulate PPARα and MCPT-1, resulting in the reduction of fatty acid oxidation activity[135-138], though some contradictory results were also reported[139,140]. Recently, core protein is reported to induce the expression of miRNA 27 to repress PPARα expression[73]. Down-regulation of PPARα and MCPT-1 by core protein may be mediated by repressing the SIRT1-AMPK signaling pathway[141].
Recently, a bipartite model has been proposed as a novel mechanism for core protein-induced steatosis: core protein first requires DGAT1 to gain access to lipid droplets[142], and then lipid droplets-localized core protein interferes with triglyceride turnover, thus stabilizing lipid droplets and leading to steatosis[143].
In the transgenic mice, HCV core-induced nonobese hepatic steatosis is associated with down-regulation of the leptin gene in visceral fat and concurrent hypoadiponectinemia. Moreover, steatosis is ameliorated by adiponectin administration[144].
Collectively, the multiple activities of core protein may participate in the triglyceride accumulation in chronic HCV infection.
The higher prevalence and much more severity of liver steatosis are observed in patients infected with HCV genotype 3 than in patients infected with other genotypes. Indeed, core protein from genotype 3a but not from genotype 1b could down-regulate PTEN in hepatocytes and trigger the formation of large lipid droplets[145]. Recent studies examining a possible mechanism of steatosis formation in genotype 3a isolates have focused on the a.a. 164 of core protein[146]. Core protein has Phe at 164 position up-regulated fatty acid synthetase promoter stronger[147].
Sequence variations in core protein may affect steatosis development. Indeed, substitutions at a.a. position 70 and/or 91 of the genotype 1b core gene are associated with the lipid accumulation that causes steatosis[148,149]. Moreover, polymorphisms at the a.a. position 182 and 186 of the genotype 3 core gene are correlated with the intrahepatic steatosis[150].
The molecular mechanism(s) of HCV-related fibrosis is unclear. Hepatic stellate cells (HSCs), one of the sinusoid constituent cells, play a critical role in liver fibrosis[151]. Oxidative stress and various cytokines are well known profibrogenic mediators[152]. HCV may induce fibrosis by the following mechanisms: (1) stimulating secretion of profibrogenic cytokines in hepatocytes; (2) interacting with sinusoidal endothelium; and (3) provoking fibrogenesis via HSCs. Transgenic mice with conditional core protein expression developed inflammation and fibrosis[52]. Therefore, core protein plays an important role in liver fibrosis.
Core protein could stimulate secretion of profibrogenic cytokines in hepatocytes. Indeed, core protein is known to up-regulate TGFβ1 expression in hepatoma cells[153]. Moreover, core protein in hepatoma cells promotes liver fibrogenesis via up-regulation of CTGF with TGFβ1 when co-cultured with HSCs[154]. Recently, TGF-β was also reported to be up-regulated in transgenic mice with core protein. Moreover, hepatoma cells expressing core protein could activate stellate cells in the co-culture system and this activation was TGF-β dependent[155]. In addition, core protein could increase the TNF-α production in monocytes[72].
Core protein could also provoke fibrogenesis via HSCs directly. Non-enveloped core protein could be secreted by infected cells[156,157]. These secreted core proteins could stimulate fibrosis in HSCs via either obese receptor[158] or toll-like receptor 2[159].
Little is known regarding the molecular mechanism(s) of HCV-related cirrhosis. Core protein could up-regulate and sustain HIF-1α expression under hypoxia, thereby contributing to increased VEGF expression, a key regulator in the hypoxic milieu of liver cirrhosis[160]. Therefore, core protein may play a role in liver cirrhosis through the up-regulation of VEGF expression.
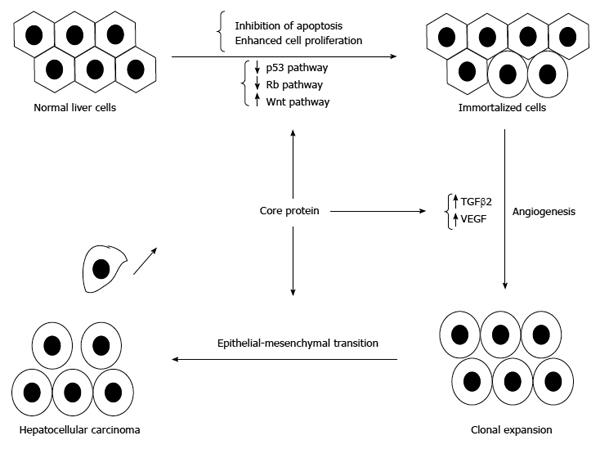
The major risk factor for the development of HCC in HCV-infected patients is pre-existing cirrhosis. Therefore, the main hypothesis for HCV carcinogenesis is that it occurs through the effects of chronic inflammation and hepatocellular injury. However, HCC can still develop in a small proportion of non-cirrhotic patients with chronic hepatitis C, suggesting that HCV may be directly involved in hepatocarcinogenesis. This was supported by the report that transgenic mice with constitutive expression of HCV structural and nonstructural proteins would develop HCC[161]. HCV core protein plays a very important role in the development of HCC. This was supported by the report that transgenic mice with constitutive HCV core protein expression would develop HCC[162].
Core protein may induce HCC development through its contribution to the onset of oxidative stress, steatosis and anti-apoptosis[48,50,76]. In addition to overcoming apoptosis, disruption of hepatocyte growth control is another key molecular event leading to the development of HCC. Actually, core protein could increase cell proliferation through the interaction with cellular proteins (e.g., p53, p73 and pRb), or through the modulation of cellular gene expressions (e.g., p21) and intracellular signal transductions, such as MAPK and Wnt/β-catenin pathways[56-58,163,164]. Moreover, core protein could stimulate primary human hepatocytes to escape from senescence and promote an immortalized phenotype[165,166] (Figure 6).
Furthermore, core protein is reported to trigger hepatic angiogenesis by induction of TGF-β2 and VEGF[167].
Core proteins derived from HCC were demonstrated to shift the TGF-β responses from tumor suppression to epithelial-mesenchymal transition (EMT)[168]. Recently, core protein is reported to epigenetically silence SFRP1 and enhance HCC aggressiveness by inducing EMT[169]. Another report also showed that core protein could promote the migration and invasion of hepatocyte via activating transcription of extracellular matrix metalloproteinase inducer[170].
Collectively, the multiple regulatory activities of core protein may result in the development of HCC.
HCV core protein-induced pathogenesis may be genotype-specific. Indeed, over expression of core gene from genotype 3a showed stronger effect in regulating expression of Cox-2 as compared to that from genotype 1a in Huh-7 cells[171]. Sequence variations within HCV core gene are reported in tumors and adjacent non-tumor tissues from the same patients[172-174]. Therefore, sequence variations of core protein within the same genotype may have different pathogenic effect. Indeed, amino acid substitutions in the core protein of genotype 1b are associated with HCC development[175-178], especially at amino acids 70 and 91[84].
In addition to HCC, HCV core protein may be also associated with the development of intrahepatic cholangiocarcinoma (ICC) and hilar cholangiocarcinoma[70,179]. However, more studies are needed to reveal the molecular mechanisms.
When co-infection with HIV/HCV occurs, HCC is more likely to occur at a younger age and with a shorter duration of HCV infection compared to those with HCV mono-infection[180]. This indicates HIV could worsen the pathogenic effects caused by HCV. On the other hand, HCV core protein could interact with HIV-1 Nef protein to stimulate HIV-1 replication in macrophages[181]. Moreover, HCV core protein could induce neuroimmune activation and potentiate HIV-1 neurotoxicity[182].
Dual infection with HCV and HBV in cirrhotic patients has been linked to an increased risk of HCC[183], indicated the interactions between these two viruses. A zebrafish model of ICC was established recently by dual expression of hepatitis B virus X protein (HBx) and HCV core protein in liver. Further studies in this model revealed that TGF-beta1 plays an important role in HBx- and HCV core protein-induced ICC development[184].
Though the exact C terminus of core protein has not been determined yet, it is likely that the mature core protein contains 177 amino acid residues[15,19]. Therefore, the cleaved peptide generated by the cleavages of both SP and SPP is from amino acid 178 to 191 (Figure 1A). This peptide is the E1 signal peptide region that facilitates the proper cleavage at core-E1 junction. All signal peptide sequences contain a hydrophobic core region, but, despite of this, they show great variation in both overall length and amino acid sequence[185]. However, the cleaved peptide (a.a. 178-191) is highly conserved with close to 100% identity among different HCV genotypes[19,29]. Sequence conservation in this cleaved peptide suggests that it should play important roles during virus infection. However, no individual residue among these 14 amino acids of the cleaved peptide is absolutely required for infectious virus production, as individual substitutions resulted in wild-type titers and a core protein fragment comprising residues 1 to 177 efficiently complemented assembly in trans[19]. Signal peptides have been suggested to have additional functions[186]. For example, the signal peptide of the lymphocytic choriomeningitis virus glycoprotein is presented by major histocompatibility complex class I as an immunodominant epitope[187], and the liberated HIV-1 gp160 leader sequence is associated with calmodulin[185,186]. A previous report argued for an additional function for this cleaved peptide. The synthetic peptide containing HCV core protein a.a. 178-187, which shows sequence homology with CYP2A6 and CYP2A7, could induce primary CTL responses in peripheral blood mononuclear cells in an HLA-A*0201-restricted manner[188]. If the cleaved peptide (a.a. 178-191) was further processed into (a.a. 178-187), it will be interesting to know how this process occurs in the cells.
Amino acid substitutions at positions 182 and 186 of the HCV genotype 3a core protein have been identified to cause increased intracellular lipid accumulation in hepatic cells[150]. These amino acid substitutions did not affect the production of mature core protein[150], in agreement with the results of a previous report[19]. Therefore, polymorphisms in the cleaved peptide (a.a. 178-191) may contribute to steatosis development. Jhaveri et al[150]. speculated (1) that the cleaved peptide interacts with host proteins within the ER membrane that mediate lipid metabolism and trafficking; and (2) that this interaction may differ between genotypes. It will be interesting to find out which cellular proteins could interact with this cleaved peptide.
Detection of the specific antibodies against ARF/core+1 proteins and the T-cell responses in HCV-infected patients provided strong evidence that ARF/core+1 protein is expressed in vivo[30,32,33,189]. However, abolishing the production of ARF/core+1 proteins had no effect on HCV replication in cultured cells or uPA-SCID mice, suggesting that ARF/core+1 proteins is probably not important for the HCV reproductive cycle[190]. On the other hand, the gene sequence conservation of this open reading frame argues that ARF/core+1 proteins should serve an important function[34].
The role of ARF/core+1 proteins in viral pathogenesis is largely unknown. It was shown that the F protein, unlike core protein, is not involved in NF-kappaB regulation[191]. Moreover, F protein, unlike core protein, could not up-regulate the expression of the fibrosis marker α-smooth muscle actin[58]. Actually, F protein does not share the major properties identified previously for the core protein, other than repressing p21 expression[192]. Down-regulation of p21 expression by F protein suggests that F protein may regulate cellular proliferation. Cellular MM-1 protein was found to interact with F protein. Further analysis indicated that F protein can enhance the gene trans-activation activity of c-Myc, apparently by antagonizing the inhibitory effect of MM-1[193]. The ability of F protein to enhance the activity of c-Myc also raises the possibility that F protein may enhance cellular proliferation. Indeed, F protein could induce hepatocyte proliferation in the transgenic mice possibly through β-catenin signaling pathway[58]. These results suggest that F protein may play a role in hepatocellular transformation in HCV patients. This hypothesis was supported by the finding that HCV sequences derived from HCC tissues could produce F protein more efficiently than those derived from non-HCC tissues[173,174,194]. Moreover, high occurrence of anti-core+1 antibodies was detected in the serum of HCC patients[195].
HCV core protein may regulate iron metabolism through interacting with FTL[54]. Recently, core+1/ARF protein was found to decrease hepcidin transcription through an AP1 binding site[196]. This indicates that core+1/ARF protein may also affect iron metabolism because hepcidin is the main regulator of iron metabolism. Moreover, HCV core and F proteins were shown to induce hepatocyte proliferation in the transgenic mice possibly through β-catenin signaling pathway[58]. Therefore, these two proteins seem to have redundant pathogenic roles. This explains the findings that HCV patients who do not produce normal anti-core antibodies have unusually high levels of anti-core+1/ARFP[197], and that the HCV-1b core+1 products are negatively regulated by core expression[38].
F protein was also found to interact with cellular prefoldin 2 protein. Moreover, expression of F protein resulted in aberrant organization of tubulin cytoskeleton[198], which suggests that F protein may affect cellular functions. On the other hand, it is possible that F protein may serve as a modulator to prevent high level of HCV replication and thus contributes to viral persistence in chronic HCV infection since HCV replication requires intact microtubule and actin polymerization[198].
It is not known whether different ARF/core+1 proteins regulate cellular activities through similar pathways. Similar subcellular localization and short half-lives of F and core+1/S proteins[41,42] suggest that these two proteins may have similar regulatory activities. However, further investigations are needed to clarify this issue.
In addition to the mature core protein (a.a. 1-177) and the cleaved peptide (a.a. 178-191) encoded from the conventional open reading frame of HCV core gene, several ARF/core+1 proteins could be expressed from the core+1 reading frame.
Through interacting with cellular proteins, modulating cellular gene expression, inducing reactive oxygen species production, and modulating cellular apoptosis, core protein could induce insulin resistance, steatosis, and even hepatocellular carcinoma. The cleaved peptide (a.a. 178-191) may play a role in the immune response and steatosis development. Though labile, ARFP/core+1/F protein could interact with cellular proteins and enhance hepatocyte growth. The core+1/ARF protein may also affect iron metabolism.
The present-day knowledge about the pathogenic roles of core gene products discussed here is obtained from cell culture and transgenic mouse models. The transgenic mice with constitutive expression of core gene products are tolerant to these proteins, leading to an insufficient immune response. A Cre/loxP recombination system has been developed in transgenic mice to study the inflammation caused by core protein. This inducible system in transgenic mice may be suitable to study the fibrosis and cirrhosis caused by core protein in the near future.
Studies of the cellular mechanisms involved in the pathogenesis of core gene products should help in the design of therapeutic drugs.
P- Reviewers: Iwasaki Y, Sazci A S- Editor: Gou SX L- Editor: A E- Editor: Zhang DN
| 1. | Timm J, Roggendorf M. Sequence diversity of hepatitis C virus: implications for immune control and therapy. World J Gastroenterol. 2007;13:4808-4817. [PubMed] [Cited in This Article: ] |
| 2. | Alberti A, Chemello L, Benvegnù L. Natural history of hepatitis C. J Hepatol. 1999;31 Suppl 1:17-24. [PubMed] [Cited in This Article: ] |
| 3. | Hoofnagle JH. Course and outcome of hepatitis C. Hepatology. 2002;36:S21-S29. [PubMed] [Cited in This Article: ] |
| 4. | Chen SL, Morgan TR. The natural history of hepatitis C virus (HCV) infection. Int J Med Sci. 2006;3:47-52. [PubMed] [Cited in This Article: ] |
| 5. | Grimbert S, Valensi P, Lévy-Marchal C, Perret G, Richardet JP, Raffoux C, Trinchet JC, Beaugrand M. High prevalence of diabetes mellitus in patients with chronic hepatitis C. A case-control study. Gastroenterol Clin Biol. 1996;20:544-548. [PubMed] [Cited in This Article: ] |
| 6. | Montenegro L, De Michina A, Misciagna G, Guerra V, Di Leo A. Virus C hepatitis and type 2 diabetes: a cohort study in southern Italy. Am J Gastroenterol. 2013;108:1108-1111. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 42] [Cited by in F6Publishing: 39] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 7. | Penin F, Dubuisson J, Rey FA, Moradpour D, Pawlotsky JM. Structural biology of hepatitis C virus. Hepatology. 2004;39:5-19. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 458] [Cited by in F6Publishing: 446] [Article Influence: 22.3] [Reference Citation Analysis (0)] |
| 8. | Lemon SM, Walker CM, Alter MJ, Yi M. Hepatitis C Virus. Fields' Virology. 5th ed. Philadelphia: Lippincott Williams and Wilkins 2007; 1253-1304. [Cited in This Article: ] |
| 9. | Kato N, Hijikata M, Ootsuyama Y, Nakagawa M, Ohkoshi S, Shimotohno K. Sequence diversity of hepatitis C viral genomes. Mol Biol Med. 1990;7:495-501. [PubMed] [Cited in This Article: ] |
| 10. | Gómez J, Martell M, Quer J, Cabot B, Esteban JI. Hepatitis C viral quasispecies. J Viral Hepat. 1999;6:3-16. [PubMed] [Cited in This Article: ] |
| 11. | Chayama K, Hayes CN. Hepatitis C virus: How genetic variability affects pathobiology of disease. J Gastroenterol Hepatol. 2011;26 Suppl 1:83-95. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 48] [Cited by in F6Publishing: 47] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 12. | Bukh J, Miller RH, Purcell RH. Genetic heterogeneity of hepatitis C virus: quasispecies and genotypes. Semin Liver Dis. 1995;15:41-63. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 574] [Cited by in F6Publishing: 589] [Article Influence: 20.3] [Reference Citation Analysis (1)] |
| 13. | Simmonds P. Genetic diversity and evolution of hepatitis C virus--15 years on. J Gen Virol. 2004;85:3173-3188. [PubMed] [Cited in This Article: ] |
| 14. | Santolini E, Migliaccio G, La Monica N. Biosynthesis and biochemical properties of the hepatitis C virus core protein. J Virol. 1994;68:3631-3641. [PubMed] [Cited in This Article: ] |
| 15. | Okamoto K, Mori Y, Komoda Y, Okamoto T, Okochi M, Takeda M, Suzuki T, Moriishi K, Matsuura Y. Intramembrane processing by signal peptide peptidase regulates the membrane localization of hepatitis C virus core protein and viral propagation. J Virol. 2008;82:8349-8361. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 82] [Cited by in F6Publishing: 90] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 16. | Ma HC, Ku YY, Hsieh YC, Lo SY. Characterization of the cleavage of signal peptide at the C-terminus of hepatitis C virus core protein by signal peptide peptidase. J Biomed Sci. 2007;14:31-41. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 19] [Cited by in F6Publishing: 21] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 17. | Ogino T, Fukuda H, Imajoh-Ohmi S, Kohara M, Nomoto A. Membrane binding properties and terminal residues of the mature hepatitis C virus capsid protein in insect cells. J Virol. 2004;78:11766-11777. [PubMed] [Cited in This Article: ] |
| 18. | Hüssy P, Faust H, Wagner JC, Schmid G, Mous J, Jacobsen H. Evaluation of hepatitis C virus envelope proteins expressed in E. coli and insect cells for use as tools for antibody screening. J Hepatol. 1997;26:1179-1186. [PubMed] [Cited in This Article: ] |
| 19. | Kopp M, Murray CL, Jones CT, Rice CM. Genetic analysis of the carboxy-terminal region of the hepatitis C virus core protein. J Virol. 2010;84:1666-1673. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 33] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 20. | Davis GL. Hepatitis C virus genotypes and quasispecies. Am J Med. 1999;107:21S-26S. [PubMed] [Cited in This Article: ] |
| 21. | Simmonds P, Smith DB, McOmish F, Yap PL, Kolberg J, Urdea MS, Holmes EC. Identification of genotypes of hepatitis C virus by sequence comparisons in the core, E1 and NS-5 regions. J Gen Virol. 1994;75:1053-1061. [PubMed] [Cited in This Article: ] |
| 22. | Kota S, Takahashi V, Ni F, Snyder JK, Strosberg AD. Direct binding of a hepatitis C virus inhibitor to the viral capsid protein. PLoS One. 2012;7:e32207. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 19] [Cited by in F6Publishing: 20] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 23. | Fukasawa M. Cellular lipid droplets and hepatitis C virus life cycle. Biol Pharm Bull. 2010;33:355-359. [PubMed] [Cited in This Article: ] |
| 24. | Mori Y, Moriishi K, Matsuura Y. Hepatitis C virus core protein: its coordinate roles with PA28gamma in metabolic abnormality and carcinogenicity in the liver. Int J Biochem Cell Biol. 2008;40:1437-1442. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 23] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 25. | Boulant S, Montserret R, Hope RG, Ratinier M, Targett-Adams P, Lavergne JP, Penin F, McLauchlan J. Structural determinants that target the hepatitis C virus core protein to lipid droplets. J Biol Chem. 2006;281:22236-22247. [PubMed] [Cited in This Article: ] |
| 26. | Suzuki R, Sakamoto S, Tsutsumi T, Rikimaru A, Tanaka K, Shimoike T, Moriishi K, Iwasaki T, Mizumoto K, Matsuura Y. Molecular determinants for subcellular localization of hepatitis C virus core protein. J Virol. 2005;79:1271-1281. [PubMed] [Cited in This Article: ] |
| 27. | Lo SY, Masiarz F, Hwang SB, Lai MM, Ou JH. Differential subcellular localization of hepatitis C virus core gene products. Virology. 1995;213:455-461. [PubMed] [Cited in This Article: ] |
| 28. | Moriishi K, Okabayashi T, Nakai K, Moriya K, Koike K, Murata S, Chiba T, Tanaka K, Suzuki R, Suzuki T. Proteasome activator PA28gamma-dependent nuclear retention and degradation of hepatitis C virus core protein. J Virol. 2003;77:10237-10249. [PubMed] [Cited in This Article: ] |
| 29. | Bukh J, Purcell RH, Miller RH. Sequence analysis of the core gene of 14 hepatitis C virus genotypes. Proc Natl Acad Sci USA. 1994;91:8239-8243. [PubMed] [Cited in This Article: ] |
| 30. | Walewski JL, Keller TR, Stump DD, Branch AD. Evidence for a new hepatitis C virus antigen encoded in an overlapping reading frame. RNA. 2001;7:710-721. [PubMed] [Cited in This Article: ] |
| 31. | Lo SY, Selby M, Tong M, Ou JH. Comparative studies of the core gene products of two different hepatitis C virus isolates: two alternative forms determined by a single amino acid substitution. Virology. 1994;199:124-131. [PubMed] [Cited in This Article: ] |
| 32. | Xu Z, Choi J, Yen TS, Lu W, Strohecker A, Govindarajan S, Chien D, Selby MJ, Ou J. Synthesis of a novel hepatitis C virus protein by ribosomal frameshift. EMBO J. 2001;20:3840-3848. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 211] [Cited by in F6Publishing: 218] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 33. | Varaklioti A, Vassilaki N, Georgopoulou U, Mavromara P. Alternate translation occurs within the core coding region of the hepatitis C viral genome. J Biol Chem. 2002;277:17713-17721. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 108] [Cited by in F6Publishing: 113] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 34. | Branch AD, Stump DD, Gutierrez JA, Eng F, Walewski JL. The hepatitis C virus alternate reading frame (ARF) and its family of novel products: the alternate reading frame protein/F-protein, the double-frameshift protein, and others. Semin Liver Dis. 2005;25:105-117. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 94] [Cited by in F6Publishing: 99] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 35. | Boulant S, Becchi M, Penin F, Lavergne JP. Unusual multiple recoding events leading to alternative forms of hepatitis C virus core protein from genotype 1b. J Biol Chem. 2003;278:45785-45792. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 62] [Cited by in F6Publishing: 67] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 36. | Baril M, Brakier-Gingras L. Translation of the F protein of hepatitis C virus is initiated at a non-AUG codon in a +1 reading frame relative to the polyprotein. Nucleic Acids Res. 2005;33:1474-1486. [PubMed] [Cited in This Article: ] |
| 37. | Vassilaki N, Boleti H, Mavromara P. Expression studies of the HCV-1a core+1 open reading frame in mammalian cells. Virus Res. 2008;133:123-135. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 21] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 38. | Boumlic A, Vassilaki N, Dalagiorgou G, Kochlios E, Kakkanas A, Georgopoulou U, Markoulatos P, Orfanoudakis G, Mavromara P. Internal translation initiation stimulates expression of the ARF/core+1 open reading frame of HCV genotype 1b. Virus Res. 2011;155:213-220. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 13] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 39. | Wolf M, Dimitrova M, Baumert TF, Schuster C. The major form of hepatitis C virus alternate reading frame protein is suppressed by core protein expression. Nucleic Acids Res. 2008;36:3054-3064. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 21] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 40. | Vassilaki N, Mavromara P. The HCV ARFP/F/core+1 protein: production and functional analysis of an unconventional viral product. IUBMB Life. 2009;61:739-752. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 38] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 41. | Vassilaki N, Boleti H, Mavromara P. Expression studies of the core+1 protein of the hepatitis C virus 1a in mammalian cells. The influence of the core protein and proteasomes on the intracellular levels of core+1. FEBS J. 2007;274:4057-4074. [PubMed] [Cited in This Article: ] |
| 42. | Xu Z, Choi J, Lu W, Ou JH. Hepatitis C virus f protein is a short-lived protein associated with the endoplasmic reticulum. J Virol. 2003;77:1578-1583. [PubMed] [Cited in This Article: ] |
| 43. | Yuksek K, Chen WL, Chien D, Ou JH. Ubiquitin-independent degradation of hepatitis C virus F protein. J Virol. 2009;83:612-621. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 45] [Cited by in F6Publishing: 47] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 44. | Boumlic A, Nominé Y, Charbonnier S, Dalagiorgou G, Vassilaki N, Kieffer B, Travé G, Mavromara P, Orfanoudakis G. Prevalence of intrinsic disorder in the hepatitis C virus ARFP/Core+1/S protein. FEBS J. 2010;277:774-789. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 13] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 45. | Kremsdorf D, Brezillon N. New animal models for hepatitis C viral infection and pathogenesis studies. World J Gastroenterol. 2007;13:2427-2435. [PubMed] [Cited in This Article: ] |
| 46. | Fujinaga H, Tsutsumi T, Yotsuyanagi H, Moriya K, Koike K. Hepatocarcinogenesis in hepatitis C: HCV shrewdly exacerbates oxidative stress by modulating both production and scavenging of reactive oxygen species. Oncology. 2011;81 Suppl 1:11-17. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 46] [Cited by in F6Publishing: 47] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 47. | Chang ML, Chen JC, Yeh CT, Sheen IS, Tai DI, Chang MY, Chiu CT, Lin DY, Bissell DM. Topological and evolutional relationships between HCV core protein and hepatic lipid vesicles: studies in vitro and in conditionally transgenic mice. World J Gastroenterol. 2007;13:3472-3477. [PubMed] [Cited in This Article: ] |
| 48. | Moriya K, Nakagawa K, Santa T, Shintani Y, Fujie H, Miyoshi H, Tsutsumi T, Miyazawa T, Ishibashi K, Horie T. Oxidative stress in the absence of inflammation in a mouse model for hepatitis C virus-associated hepatocarcinogenesis. Cancer Res. 2001;61:4365-4370. [PubMed] [Cited in This Article: ] |
| 49. | Miyamoto H, Moriishi K, Moriya K, Murata S, Tanaka K, Suzuki T, Miyamura T, Koike K, Matsuura Y. Involvement of the PA28gamma-dependent pathway in insulin resistance induced by hepatitis C virus core protein. J Virol. 2007;81:1727-1735. [PubMed] [Cited in This Article: ] |
| 50. | Moriishi K, Mochizuki R, Moriya K, Miyamoto H, Mori Y, Abe T, Murata S, Tanaka K, Miyamura T, Suzuki T. Critical role of PA28gamma in hepatitis C virus-associated steatogenesis and hepatocarcinogenesis. Proc Natl Acad Sci USA. 2007;104:1661-1666. [PubMed] [Cited in This Article: ] |
| 51. | Shintani Y, Fujie H, Miyoshi H, Tsutsumi T, Tsukamoto K, Kimura S, Moriya K, Koike K. Hepatitis C virus infection and diabetes: direct involvement of the virus in the development of insulin resistance. Gastroenterology. 2004;126:840-848. [PubMed] [Cited in This Article: ] |
| 52. | Chang ML, Yeh CT, Lin DY, Ho YP, Hsu CM, Bissell DM. Hepatic inflammation mediated by hepatitis C virus core protein is ameliorated by blocking complement activation. BMC Med Genomics. 2009;2:51. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 15] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 53. | de Chassey B, Navratil V, Tafforeau L, Hiet MS, Aublin-Gex A, Agaugué S, Meiffren G, Pradezynski F, Faria BF, Chantier T. Hepatitis C virus infection protein network. Mol Syst Biol. 2008;4:230. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 295] [Cited by in F6Publishing: 295] [Article Influence: 18.4] [Reference Citation Analysis (0)] |
| 54. | Tripathi LP, Kataoka C, Taguwa S, Moriishi K, Mori Y, Matsuura Y, Mizuguchi K. Network based analysis of hepatitis C virus core and NS4B protein interactions. Mol Biosyst. 2010;6:2539-2553. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 41] [Cited by in F6Publishing: 44] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 55. | Ivanyi-Nagy R, Darlix JL. Fuzziness in the core of the human pathogenic viruses HCV and HIV. Adv Exp Med Biol. 2012;725:142-158. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 8] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 56. | Banerjee A, Ray RB, Ray R. Oncogenic potential of hepatitis C virus proteins. Viruses. 2010;2:2108-2133. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 83] [Cited by in F6Publishing: 90] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 57. | Fukutomi T, Zhou Y, Kawai S, Eguchi H, Wands JR, Li J. Hepatitis C virus core protein stimulates hepatocyte growth: correlation with upregulation of wnt-1 expression. Hepatology. 2005;41:1096-1105. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 113] [Cited by in F6Publishing: 122] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 58. | Hu WT, Li HC, Lee SK, Ma HC, Yang CH, Chen HL, Lo SY. Both core and F proteins of hepatitis C virus could enhance cell proliferation in transgenic mice. Biochem Biophys Res Commun. 2013;435:147-152. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 12] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 59. | Basu A, Meyer K, Lai KK, Saito K, Di Bisceglie AM, Grosso LE, Ray RB, Ray R. Microarray analyses and molecular profiling of Stat3 signaling pathway induced by hepatitis C virus core protein in human hepatocytes. Virology. 2006;349:347-358. [PubMed] [Cited in This Article: ] |
| 60. | Pazienza V, Clément S, Pugnale P, Conzelmann S, Pascarella S, Mangia A, Negro F. Gene expression profile of Huh-7 cells expressing hepatitis C virus genotype 1b or 3a core proteins. Liver Int. 2009;29:661-669. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 24] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 61. | Chang ML, Yeh CT, Chen JC, Huang CC, Lin SM, Sheen IS, Tai DI, Chu CM, Lin WP, Chang MY. Altered expression patterns of lipid metabolism genes in an animal model of HCV core-related, nonobese, modest hepatic steatosis. BMC Genomics. 2008;9:109. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 31] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 62. | Budhu A, Chen Y, Kim JW, Forgues M, Valerie K, Harris CC, Wang XW. Induction of a unique gene expression profile in primary human hepatocytes by hepatitis C virus core, NS3 and NS5A proteins. Carcinogenesis. 2007;28:1552-1560. [PubMed] [Cited in This Article: ] |
| 63. | Jhaveri R, Kundu P, Shapiro AM, Venkatesan A, Dasgupta A. Effect of heptitis C virus core protein on cellular gene expression: specific inhibition of cyclooxygenase 2. J Infect Dis. 2005;191:1498-1506. [PubMed] [Cited in This Article: ] |
| 64. | Wu CG, Budhu A, Chen S, Zhou X, Popescu NC, Valerie K, Wang XW. Effect of hepatitis C virus core protein on the molecular profiling of human B lymphocytes. Mol Med. 2006;12:47-53. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 25] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 65. | Liu M, Zhang SL, Cheng J, Liu Y, Wang L, Shao Q, Zhang J, Lin SM. Genes transactivated by hepatitis C virus core protein, a microarray assay. World J Gastroenterol. 2005;11:3351-3356. [PubMed] [Cited in This Article: ] |
| 66. | Dou J, Liu P, Zhang X. Cellular response to gene expression profiles of different hepatitis C virus core proteins in the Huh-7 cell line with microarray analysis. J Nanosci Nanotechnol. 2005;5:1230-1235. [PubMed] [Cited in This Article: ] |
| 67. | Dou J, Liu P, Wang J, Zhang X. Preliminary analysis of gene expression profiles in HepG2 cell line induced by different genotype core proteins of HCV. Cell Mol Immunol. 2006;3:227-233. [PubMed] [Cited in This Article: ] |
| 68. | Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215-233. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14460] [Cited by in F6Publishing: 15413] [Article Influence: 1027.5] [Reference Citation Analysis (1)] |
| 69. | Li S, Xing X, Yang Q, Xu H, He J, Chen Z, Zhu H. The effects of hepatitis C virus core protein on the expression of miR-122 in vitro. Virol J. 2013;10:98. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 10] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 70. | Zeng B, Li Z, Chen R, Guo N, Zhou J, Zhou Q, Lin Q, Cheng D, Liao Q, Zheng L. Epigenetic regulation of miR-124 by hepatitis C virus core protein promotes migration and invasion of intrahepatic cholangiocarcinoma cells by targeting SMYD3. FEBS Lett. 2012;586:3271-3278. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 77] [Cited by in F6Publishing: 77] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 71. | Shiu TY, Huang SM, Shih YL, Chu HC, Chang WK, Hsieh TY. Hepatitis C virus core protein down-regulates p21(Waf1/Cip1) and inhibits curcumin-induced apoptosis through microRNA-345 targeting in human hepatoma cells. PLoS One. 2013;8:e61089. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 35] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 72. | Bala S, Tilahun Y, Taha O, Alao H, Kodys K, Catalano D, Szabo G. Increased microRNA-155 expression in the serum and peripheral monocytes in chronic HCV infection. J Transl Med. 2012;10:151. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 120] [Cited by in F6Publishing: 126] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 73. | Singaravelu R, Chen R, Lyn RK, Jones DM, O’Hara S, Rouleau Y, Cheng J, Srinivasan P, Nasheri N, Russell RS. Hepatitis C virus induced up-regulation of microRNA-27: a novel mechanism for hepatic steatosis. Hepatology. 2014;59:98-108. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 80] [Cited by in F6Publishing: 84] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 74. | Gu Y, Xu Y, Jiang L, Cao X, Liu F, Li H, Zhang L, Li Z, Li J, Ye J. Differentially expressed microRNAs in Huh-7 cells expressing HCV core genotypes 3a or 1b: potential functions and downstream pathways. Int J Mol Med. 2012;30:374-382. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 8] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 75. | Fischer R, Baumert T, Blum HE. Hepatitis C virus infection and apoptosis. World J Gastroenterol. 2007;13:4865-4872. [PubMed] [Cited in This Article: ] |
| 76. | Jahan S, Ashfaq UA, Khaliq S, Samreen B, Afzal N. Dual behavior of HCV Core gene in regulation of apoptosis is important in progression of HCC. Infect Genet Evol. 2012;12:236-239. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 29] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 77. | Ruggieri A, Harada T, Matsuura Y, Miyamura T. Sensitization to Fas-mediated apoptosis by hepatitis C virus core protein. Virology. 1997;229:68-76. [PubMed] [Cited in This Article: ] |
| 78. | Machida K, Tsukiyama-Kohara K, Seike E, Toné S, Shibasaki F, Shimizu M, Takahashi H, Hayashi Y, Funata N, Taya C. Inhibition of cytochrome c release in Fas-mediated signaling pathway in transgenic mice induced to express hepatitis C viral proteins. J Biol Chem. 2001;276:12140-12146. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 80] [Cited by in F6Publishing: 86] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 79. | Ray RB, Meyer K, Steele R, Shrivastava A, Aggarwal BB, Ray R. Inhibition of tumor necrosis factor (TNF-alpha)-mediated apoptosis by hepatitis C virus core protein. J Biol Chem. 1998;273:2256-2259. [PubMed] [Cited in This Article: ] |
| 80. | Zhu N, Khoshnan A, Schneider R, Matsumoto M, Dennert G, Ware C, Lai MM. Hepatitis C virus core protein binds to the cytoplasmic domain of tumor necrosis factor (TNF) receptor 1 and enhances TNF-induced apoptosis. J Virol. 1998;72:3691-3697. [PubMed] [Cited in This Article: ] |
| 81. | Saito K, Meyer K, Warner R, Basu A, Ray RB, Ray R. Hepatitis C virus core protein inhibits tumor necrosis factor alpha-mediated apoptosis by a protective effect involving cellular FLICE inhibitory protein. J Virol. 2006;80:4372-4379. [PubMed] [Cited in This Article: ] |
| 82. | Jahan S, Khaliq S, Siddiqi MH, Ijaz B, Ahmad W, Ashfaq UA, Hassan S. Anti-apoptotic effect of HCV core gene of genotype 3a in Huh-7 cell line. Virol J. 2011;8:522. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 15] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 83. | Pavio N, Lai MM. The hepatitis C virus persistence: how to evade the immune system? J Biosci. 2003;28:287-304. [PubMed] [Cited in This Article: ] |
| 84. | Khaliq S, Jahan S, Pervaiz A. Sequence variability of HCV Core region: important predictors of HCV induced pathogenesis and viral production. Infect Genet Evol. 2011;11:543-556. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 33] [Article Influence: 2.5] [Reference Citation Analysis (2)] |
| 85. | Anjum S, Afzal MS, Ahmad T, Aslam B, Waheed Y, Shafi T, Qadri I. Mutations in the STAT1-interacting domain of the hepatitis C virus core protein modulate the response to antiviral therapy. Mol Med Rep. 2013;8:487-492. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 9] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 86. | Lin W, Kim SS, Yeung E, Kamegaya Y, Blackard JT, Kim KA, Holtzman MJ, Chung RT. Hepatitis C virus core protein blocks interferon signaling by interaction with the STAT1 SH2 domain. J Virol. 2006;80:9226-9235. [PubMed] [Cited in This Article: ] |
| 87. | Inoue K, Tsukiyama-Kohara K, Matsuda C, Yoneyama M, Fujita T, Kuge S, Yoshiba M, Kohara M. Impairment of interferon regulatory factor-3 activation by hepatitis C virus core protein basic amino acid region 1. Biochem Biophys Res Commun. 2012;428:494-499. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 13] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 88. | Kang JI, Kwon YC, Ahn BY. Modulation of the type I interferon pathways by culture-adaptive hepatitis C virus core mutants. FEBS Lett. 2012;586:1272-1278. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 12] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 89. | Oshiumi H, Ikeda M, Matsumoto M, Watanabe A, Takeuchi O, Akira S, Kato N, Shimotohno K, Seya T. Hepatitis C virus core protein abrogates the DDX3 function that enhances IPS-1-mediated IFN-beta induction. PLoS One. 2010;5:e14258. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 65] [Cited by in F6Publishing: 65] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 90. | Dominguez-Villar M, Garcia-Cozar FJ, Chambers BJ. The effects of hepatitis C virus core protein on functional responses in the NK cell line YTS. Scand J Immunol. 2012;75:54-60. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 6] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 91. | Imran M, Waheed Y, Manzoor S, Bilal M, Ashraf W, Ali M, Ashraf M. Interaction of Hepatitis C virus proteins with pattern recognition receptors. Virol J. 2012;9:126. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 44] [Cited by in F6Publishing: 43] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 92. | Mazumdar B, Kim H, Meyer K, Bose SK, Di Bisceglie AM, Ray RB, Diamond MS, Atkinson JP, Ray R. Hepatitis C virus infection upregulates CD55 expression on the hepatocyte surface and promotes association with virus particles. J Virol. 2013;87:7902-7910. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 26] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 93. | Kim H, Meyer K, Di Bisceglie AM, Ray R. Hepatitis C virus suppresses C9 complement synthesis and impairs membrane attack complex function. J Virol. 2013;87:5858-5867. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 35] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 94. | Soguero C, Joo M, Chianese-Bullock KA, Nguyen DT, Tung K, Hahn YS. Hepatitis C virus core protein leads to immune suppression and liver damage in a transgenic murine model. J Virol. 2002;76:9345-9354. [PubMed] [Cited in This Article: ] |
| 95. | Frazier AD, Zhang CL, Ni L, Ma CJ, Zhang Y, Wu XY, Atia AN, Yao ZQ, Moorman JP. Programmed death-1 affects suppressor of cytokine signaling-1 expression in T cells during hepatitis C infection. Viral Immunol. 2010;23:487-495. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 26] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 96. | Zhang Y, Ma CJ, Ni L, Zhang CL, Wu XY, Kumaraguru U, Li CF, Moorman JP, Yao ZQ. Cross-talk between programmed death-1 and suppressor of cytokine signaling-1 in inhibition of IL-12 production by monocytes/macrophages in hepatitis C virus infection. J Immunol. 2011;186:3093-3103. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 64] [Cited by in F6Publishing: 73] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 97. | Zhang Y, Ma CJ, Wang JM, Ji XJ, Wu XY, Jia ZS, Moorman JP, Yao ZQ. Tim-3 negatively regulates IL-12 expression by monocytes in HCV infection. PLoS One. 2011;6:e19664. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 66] [Cited by in F6Publishing: 79] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 98. | Tacke RS, Tosello-Trampont A, Nguyen V, Mullins DW, Hahn YS. Extracellular hepatitis C virus core protein activates STAT3 in human monocytes/macrophages/dendritic cells via an IL-6 autocrine pathway. J Biol Chem. 2011;286:10847-10855. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 72] [Cited by in F6Publishing: 75] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 99. | Kim H, Mazumdar B, Bose SK, Meyer K, Di Bisceglie AM, Hoft DF, Ray R. Hepatitis C virus-mediated inhibition of cathepsin S increases invariant-chain expression on hepatocyte surface. J Virol. 2012;86:9919-9928. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 16] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 100. | Dominguez-Villar M, Fernandez-Ponce C, Munoz-Suano A, Gomez E, Rodríguez-Iglesias M, Garcia-Cozar F. Up-regulation of FOXP3 and induction of suppressive function in CD4+ Jurkat T-cells expressing hepatitis C virus core protein. Clin Sci (Lond). 2012;123:15-27. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 17] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 101. | Akuta N, Suzuki F, Sezaki H, Suzuki Y, Hosaka T, Someya T, Kobayashi M, Saitoh S, Watahiki S, Sato J. Association of amino acid substitution pattern in core protein of hepatitis C virus genotype 1b high viral load and non-virological response to interferon-ribavirin combination therapy. Intervirology. 2005;48:372-380. [PubMed] [Cited in This Article: ] |
| 102. | Funaoka Y, Sakamoto N, Suda G, Itsui Y, Nakagawa M, Kakinuma S, Watanabe T, Mishima K, Ueyama M, Onozuka I. Analysis of interferon signaling by infectious hepatitis C virus clones with substitutions of core amino acids 70 and 91. J Virol. 2011;85:5986-5994. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 44] [Cited by in F6Publishing: 50] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 103. | Choi J, Ou JH. Mechanisms of liver injury. III. Oxidative stress in the pathogenesis of hepatitis C virus. Am J Physiol Gastrointest Liver Physiol. 2006;290:G847-G851. [PubMed] [Cited in This Article: ] |
| 104. | Korenaga M, Wang T, Li Y, Showalter LA, Chan T, Sun J, Weinman SA. Hepatitis C virus core protein inhibits mitochondrial electron transport and increases reactive oxygen species (ROS) production. J Biol Chem. 2005;280:37481-37488. [PubMed] [Cited in This Article: ] |
| 105. | Wang T, Weinman SA. Interactions Between Hepatitis C Virus and Mitochondria: Impact on Pathogenesis and Innate Immunity. Curr Pathobiol Rep. 2013;1:179-187. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in F6Publishing: 31] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 106. | Carvajal-Yepes M, Himmelsbach K, Schaedler S, Ploen D, Krause J, Ludwig L, Weiss T, Klingel K, Hildt E. Hepatitis C virus impairs the induction of cytoprotective Nrf2 target genes by delocalization of small Maf proteins. J Biol Chem. 2011;286:8941-8951. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 84] [Cited by in F6Publishing: 85] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 107. | Li K, Prow T, Lemon SM, Beard MR. Cellular response to conditional expression of hepatitis C virus core protein in Huh7 cultured human hepatoma cells. Hepatology. 2002;35:1237-1246. [PubMed] [Cited in This Article: ] |
| 108. | Ivanov AV, Smirnova OA, Ivanova ON, Masalova OV, Kochetkov SN, Isaguliants MG. Hepatitis C virus proteins activate NRF2/ARE pathway by distinct ROS-dependent and independent mechanisms in HUH7 cells. PLoS One. 2011;6:e24957. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 116] [Cited by in F6Publishing: 123] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 109. | Badar S, Khubaib B, Idrees M, Hussain A, Awan Z, Butt S, Afzal S, Akram M, Fatima Z, Aftab M. Association of hepatitis C virus with insulin resistance: evidences from animal studies and clinical studies. Hepat Mon. 2012;12:11-15. [PubMed] [Cited in This Article: ] |
| 110. | Gumber SC, Chopra S. Hepatitis C: a multifaceted disease. Review of extrahepatic manifestations. Ann Intern Med. 1995;123:615-620. [PubMed] [Cited in This Article: ] |
| 111. | Moucari R, Asselah T, Cazals-Hatem D, Voitot H, Boyer N, Ripault MP, Sobesky R, Martinot-Peignoux M, Maylin S, Nicolas-Chanoine MH. Insulin resistance in chronic hepatitis C: association with genotypes 1 and 4, serum HCV RNA level, and liver fibrosis. Gastroenterology. 2008;134:416-423. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 375] [Cited by in F6Publishing: 360] [Article Influence: 22.5] [Reference Citation Analysis (1)] |
| 112. | Del Campo JA, Romero-Gómez M. Steatosis and insulin resistance in hepatitis C: a way out for the virus? World J Gastroenterol. 2009;15:5014-5019. [PubMed] [Cited in This Article: ] |
| 113. | Serfaty L, Capeau J. Hepatitis C, insulin resistance and diabetes: clinical and pathogenic data. Liver Int. 2009;29 Suppl 2:13-25. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 63] [Cited by in F6Publishing: 65] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 114. | Nakatani Y, Kaneto H, Kawamori D, Yoshiuchi K, Hatazaki M, Matsuoka TA, Ozawa K, Ogawa S, Hori M, Yamasaki Y. Involvement of endoplasmic reticulum stress in insulin resistance and diabetes. J Biol Chem. 2005;280:847-851. [PubMed] [Cited in This Article: ] |
| 115. | Banerjee S, Saito K, Ait-Goughoulte M, Meyer K, Ray RB, Ray R. Hepatitis C virus core protein upregulates serine phosphorylation of insulin receptor substrate-1 and impairs the downstream akt/protein kinase B signaling pathway for insulin resistance. J Virol. 2008;82:2606-2612. [PubMed] [Cited in This Article: ] |
| 116. | Pazienza V, Clément S, Pugnale P, Conzelman S, Foti M, Mangia A, Negro F. The hepatitis C virus core protein of genotypes 3a and 1b downregulates insulin receptor substrate 1 through genotype-specific mechanisms. Hepatology. 2007;45:1164-1171. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 163] [Cited by in F6Publishing: 182] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 117. | Pazienza V, Vinciguerra M, Andriulli A, Mangia A. Hepatitis C virus core protein genotype 3a increases SOCS-7 expression through PPAR-{gamma} in Huh-7 cells. J Gen Virol. 2010;91:1678-1686. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 48] [Cited by in F6Publishing: 56] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 118. | Banks AS, Li J, McKeag L, Hribal ML, Kashiwada M, Accili D, Rothman PB. Deletion of SOCS7 leads to enhanced insulin action and enlarged islets of Langerhans. J Clin Invest. 2005;115:2462-2471. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 66] [Cited by in F6Publishing: 71] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 119. | Persico M, Capasso M, Persico E, Svelto M, Russo R, Spano D, Crocè L, La Mura V, Moschella F, Masutti F. Suppressor of cytokine signaling 3 (SOCS3) expression and hepatitis C virus-related chronic hepatitis: Insulin resistance and response to antiviral therapy. Hepatology. 2007;46:1009-1015. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 114] [Cited by in F6Publishing: 128] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 120. | Kawaguchi T, Yoshida T, Harada M, Hisamoto T, Nagao Y, Ide T, Taniguchi E, Kumemura H, Hanada S, Maeyama M. Hepatitis C virus down-regulates insulin receptor substrates 1 and 2 through up-regulation of suppressor of cytokine signaling 3. Am J Pathol. 2004;165:1499-1508. [PubMed] [Cited in This Article: ] |
| 121. | Pascarella S, Clément S, Guilloux K, Conzelmann S, Penin F, Negro F. Effects of hepatitis C virus on suppressor of cytokine signaling mRNA levels: comparison between different genotypes and core protein sequence analysis. J Med Virol. 2011;83:1005-1015. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 19] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 122. | Akuta N, Suzuki F, Hirakawa M, Kawamura Y, Yatsuji H, Sezaki H, Suzuki Y, Hosaka T, Kobayashi M, Kobayashi M. Amino acid substitutions in the hepatitis C virus core region of genotype 1b are the important predictor of severe insulin resistance in patients without cirrhosis and diabetes mellitus. J Med Virol. 2009;81:1032-1039. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 41] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 123. | Rubbia-Brandt L, Quadri R, Abid K, Giostra E, Malé PJ, Mentha G, Spahr L, Zarski JP, Borisch B, Hadengue A. Hepatocyte steatosis is a cytopathic effect of hepatitis C virus genotype 3. J Hepatol. 2000;33:106-115. [PubMed] [Cited in This Article: ] |
| 124. | Negro F, Sanyal AJ. Hepatitis C virus, steatosis and lipid abnormalities: clinical and pathogenic data. Liver Int. 2009;29 Suppl 2:26-37. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 120] [Cited by in F6Publishing: 131] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 125. | Negro F. Hepatitis C virus-induced steatosis: an overview. Dig Dis. 2010;28:294-299. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 32] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 126. | Roingeard P, Hourioux C. Hepatitis C virus core protein, lipid droplets and steatosis. J Viral Hepat. 2008;15:157-164. [PubMed] [Cited in This Article: ] |
| 127. | Moriya K, Yotsuyanagi H, Shintani Y, Fujie H, Ishibashi K, Matsuura Y, Miyamura T, Koike K. Hepatitis C virus core protein induces hepatic steatosis in transgenic mice. J Gen Virol. 1997;78:1527-1531. [PubMed] [Cited in This Article: ] |
| 128. | Khan M, Jahan S, Khaliq S, Ijaz B, Ahmad W, Samreen B, Hassan S. Interaction of the hepatitis C virus (HCV) core with cellular genes in the development of HCV-induced steatosis. Arch Virol. 2010;155:1735-1753. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 20] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 129. | Perlemuter G, Sabile A, Letteron P, Vona G, Topilco A, Chrétien Y, Koike K, Pessayre D, Chapman J, Barba G. Hepatitis C virus core protein inhibits microsomal triglyceride transfer protein activity and very low density lipoprotein secretion: a model of viral-related steatosis. FASEB J. 2002;16:185-194. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 419] [Cited by in F6Publishing: 445] [Article Influence: 20.2] [Reference Citation Analysis (0)] |
| 130. | Jackel-Cram C, Qiao L, Xiang Z, Brownlie R, Zhou Y, Babiuk L, Liu Q. Hepatitis C virus genotype-3a core protein enhances sterol regulatory element-binding protein-1 activity through the phosphoinositide 3-kinase-Akt-2 pathway. J Gen Virol. 2010;91:1388-1395. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 52] [Cited by in F6Publishing: 53] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 131. | Waris G, Felmlee DJ, Negro F, Siddiqui A. Hepatitis C virus induces proteolytic cleavage of sterol regulatory element binding proteins and stimulates their phosphorylation via oxidative stress. J Virol. 2007;81:8122-8130. [PubMed] [Cited in This Article: ] |
| 132. | Tsutsumi T, Suzuki T, Shimoike T, Suzuki R, Moriya K, Shintani Y, Fujie H, Matsuura Y, Koike K, Miyamura T. Interaction of hepatitis C virus core protein with retinoid X receptor alpha modulates its transcriptional activity. Hepatology. 2002;35:937-946. [PubMed] [Cited in This Article: ] |
| 133. | García-Mediavilla MV, Pisonero-Vaquero S, Lima-Cabello E, Benedicto I, Majano PL, Jorquera F, González-Gallego J, Sánchez-Campos S. Liver X receptor α-mediated regulation of lipogenesis by core and NS5A proteins contributes to HCV-induced liver steatosis and HCV replication. Lab Invest. 2012;92:1191-1202. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 42] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 134. | Miyoshi H, Moriya K, Tsutsumi T, Shinzawa S, Fujie H, Shintani Y, Fujinaga H, Goto K, Todoroki T, Suzuki T. Pathogenesis of lipid metabolism disorder in hepatitis C: polyunsaturated fatty acids counteract lipid alterations induced by the core protein. J Hepatol. 2011;54:432-438. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 37] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 135. | Cheng Y, Dharancy S, Malapel M, Desreumaux P. Hepatitis C virus infection down-regulates the expression of peroxisome proliferator-activated receptor alpha and carnitine palmitoyl acyl-CoA transferase 1A. World J Gastroenterol. 2005;11:7591-7596. [PubMed] [Cited in This Article: ] |
| 136. | Yamaguchi A, Tazuma S, Nishioka T, Ohishi W, Hyogo H, Nomura S, Chayama K. Hepatitis C virus core protein modulates fatty acid metabolism and thereby causes lipid accumulation in the liver. Dig Dis Sci. 2005;50:1361-1371. [PubMed] [Cited in This Article: ] |
| 137. | Dharancy S, Malapel M, Perlemuter G, Roskams T, Cheng Y, Dubuquoy L, Podevin P, Conti F, Canva V, Philippe D. Impaired expression of the peroxisome proliferator-activated receptor alpha during hepatitis C virus infection. Gastroenterology. 2005;128:334-342. [PubMed] [Cited in This Article: ] |
| 138. | de Gottardi A, Pazienza V, Pugnale P, Bruttin F, Rubbia-Brandt L, Juge-Aubry CE, Meier CA, Hadengue A, Negro F. Peroxisome proliferator-activated receptor-alpha and -gamma mRNA levels are reduced in chronic hepatitis C with steatosis and genotype 3 infection. Aliment Pharmacol Ther. 2006;23:107-114. [PubMed] [Cited in This Article: ] |
| 139. | Tanaka N, Moriya K, Kiyosawa K, Koike K, Gonzalez FJ, Aoyama T. PPARalpha activation is essential for HCV core protein-induced hepatic steatosis and hepatocellular carcinoma in mice. J Clin Invest. 2008;118:683-694. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 34] [Cited by in F6Publishing: 116] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 140. | Koike K. Steatosis, liver injury, and hepatocarcinogenesis in hepatitis C viral infection. J Gastroenterol. 2009;44 Suppl 19:82-88. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 41] [Cited by in F6Publishing: 44] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 141. | Yu JW, Sun LJ, Liu W, Zhao YH, Kang P, Yan BZ. Hepatitis C virus core protein induces hepatic metabolism disorders through down-regulation of the SIRT1-AMPK signaling pathway. Int J Infect Dis. 2013;17:e539-e545. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 44] [Cited by in F6Publishing: 46] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 142. | Herker E, Harris C, Hernandez C, Carpentier A, Kaehlcke K, Rosenberg AR, Farese RV, Ott M. Efficient hepatitis C virus particle formation requires diacylglycerol acyltransferase-1. Nat Med. 2010;16:1295-1298. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 267] [Cited by in F6Publishing: 253] [Article Influence: 18.1] [Reference Citation Analysis (0)] |
| 143. | Harris C, Herker E, Farese RV, Ott M. Hepatitis C virus core protein decreases lipid droplet turnover: a mechanism for core-induced steatosis. J Biol Chem. 2011;286:42615-42625. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 60] [Cited by in F6Publishing: 67] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 144. | Chang ML, Yeh HC, Tsou YK, Wang CJ, Cheng HY, Sung CM, Ho YP, Chen TH, Yeh CT. HCV core-induced nonobese hepatic steatosis is associated with hypoadiponectinemia and is ameliorated by adiponectin administration. Obesity (Silver Spring). 2012;20:1474-1480. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 11] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 145. | Clément S, Peyrou M, Sanchez-Pareja A, Bourgoin L, Ramadori P, Suter D, Vinciguerra M, Guilloux K, Pascarella S, Rubbia-Brandt L. Down-regulation of phosphatase and tensin homolog by hepatitis C virus core 3a in hepatocytes triggers the formation of large lipid droplets. Hepatology. 2011;54:38-49. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 56] [Cited by in F6Publishing: 60] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 146. | Hourioux C, Patient R, Morin A, Blanchard E, Moreau A, Trassard S, Giraudeau B, Roingeard P. The genotype 3-specific hepatitis C virus core protein residue phenylalanine 164 increases steatosis in an in vitro cellular model. Gut. 2007;56:1302-1308. [PubMed] [Cited in This Article: ] |
| 147. | Jackel-Cram C, Babiuk LA, Liu Q. Up-regulation of fatty acid synthase promoter by hepatitis C virus core protein: genotype-3a core has a stronger effect than genotype-1b core. J Hepatol. 2007;46:999-1008. [PubMed] [Cited in This Article: ] |
| 148. | Sumida Y, Kanemasa K, Hara T, Inada Y, Sakai K, Imai S, Yoshida N, Yasui K, Itoh Y, Okanoue T. Impact of amino acid substitutions in hepatitis C virus genotype 1b core region on liver steatosis and glucose tolerance in non-cirrhotic patients without overt diabetes. J Gastroenterol Hepatol. 2011;26:836-842. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 15] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 149. | Tachi Y, Katano Y, Honda T, Hayashi K, Ishigami M, Itoh A, Hirooka Y, Nakano I, Samejima Y, Goto H. Impact of amino acid substitutions in the hepatitis C virus genotype 1b core region on liver steatosis and hepatic oxidative stress in patients with chronic hepatitis C. Liver Int. 2010;30:554-559. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25] [Cited by in F6Publishing: 30] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 150. | Jhaveri R, McHutchison J, Patel K, Qiang G, Diehl AM. Specific polymorphisms in hepatitis C virus genotype 3 core protein associated with intracellular lipid accumulation. J Infect Dis. 2008;197:283-291. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 53] [Cited by in F6Publishing: 58] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 151. | Friedman SL. Hepatic fibrosis -- overview. Toxicology. 2008;254:120-129. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 248] [Cited by in F6Publishing: 258] [Article Influence: 16.1] [Reference Citation Analysis (0)] |
| 152. | Nieto N. Oxidative-stress and IL-6 mediate the fibrogenic effects of [corrected] Kupffer cells on stellate cells. Hepatology. 2006;44:1487-1501. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 169] [Cited by in F6Publishing: 185] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 153. | Taniguchi H, Kato N, Otsuka M, Goto T, Yoshida H, Shiratori Y, Omata M. Hepatitis C virus core protein upregulates transforming growth factor-beta 1 transcription. J Med Virol. 2004;72:52-59. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 99] [Cited by in F6Publishing: 106] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 154. | Shin JY, Hur W, Wang JS, Jang JW, Kim CW, Bae SH, Jang SK, Yang SH, Sung YC, Kwon OJ. HCV core protein promotes liver fibrogenesis via up-regulation of CTGF with TGF-beta1. Exp Mol Med. 2005;37:138-145. [PubMed] [Cited in This Article: ] |
| 155. | Benzoubir N, Lejamtel C, Battaglia S, Testoni B, Benassi B, Gondeau C, Perrin-Cocon L, Desterke C, Thiers V, Samuel D. HCV core-mediated activation of latent TGF-β via thrombospondin drives the crosstalk between hepatocytes and stromal environment. J Hepatol. 2013;59:1160-1168. [PubMed] [Cited in This Article: ] |
| 156. | Masalova OV, Atanadze SN, Samokhvalov EI, Petrakova NV, Kalinina TI, Smirnov VD, Khudyakov YE, Fields HA, Kushch AA. Detection of hepatitis C virus core protein circulating within different virus particle populations. J Med Virol. 1998;55:1-6. [PubMed] [Cited in This Article: ] |
| 157. | Kanto T, Hayashi N, Takehara T, Hagiwara H, Mita E, Naito M, Kasahara A, Fusamoto H, Kamada T. Buoyant density of hepatitis C virus recovered from infected hosts: two different features in sucrose equilibrium density-gradient centrifugation related to degree of liver inflammation. Hepatology. 1994;19:296-302. [PubMed] [Cited in This Article: ] |
| 158. | Wu CF, Lin YL, Huang YT. Hepatitis C virus core protein stimulates fibrogenesis in hepatic stellate cells involving the obese receptor. J Cell Biochem. 2013;114:541-550. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 19] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 159. | Coenen M, Nischalke HD, Krämer B, Langhans B, Glässner A, Schulte D, Körner C, Sauerbruch T, Nattermann J, Spengler U. Hepatitis C virus core protein induces fibrogenic actions of hepatic stellate cells via toll-like receptor 2. Lab Invest. 2011;91:1375-1382. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 37] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 160. | Abe M, Koga H, Yoshida T, Masuda H, Iwamoto H, Sakata M, Hanada S, Nakamura T, Taniguchi E, Kawaguchi T. Hepatitis C virus core protein upregulates the expression of vascular endothelial growth factor via the nuclear factor-κB/hypoxia-inducible factor-1α axis under hypoxic conditions. Hepatol Res. 2012;42:591-600. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in F6Publishing: 29] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 161. | Lerat H, Honda M, Beard MR, Loesch K, Sun J, Yang Y, Okuda M, Gosert R, Xiao SY, Weinman SA. Steatosis and liver cancer in transgenic mice expressing the structural and nonstructural proteins of hepatitis C virus. Gastroenterology. 2002;122:352-365. [PubMed] [Cited in This Article: ] |
| 162. | Moriya K, Fujie H, Shintani Y, Yotsuyanagi H, Tsutsumi T, Ishibashi K, Matsuura Y, Kimura S, Miyamura T, Koike K. The core protein of hepatitis C virus induces hepatocellular carcinoma in transgenic mice. Nat Med. 1998;4:1065-1067. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 931] [Cited by in F6Publishing: 895] [Article Influence: 34.4] [Reference Citation Analysis (0)] |
| 163. | Koike K. Molecular basis of hepatitis C virus-associated hepatocarcinogenesis: lessons from animal model studies. Clin Gastroenterol Hepatol. 2005;3:S132-S135. [PubMed] [Cited in This Article: ] |
| 164. | Liu J, Ding X, Tang J, Cao Y, Hu P, Zhou F, Shan X, Cai X, Chen Q, Ling N. Enhancement of canonical Wnt/β-catenin signaling activity by HCV core protein promotes cell growth of hepatocellular carcinoma cells. PLoS One. 2011;6:e27496. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 88] [Cited by in F6Publishing: 93] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 165. | Ray RB, Meyer K, Ray R. Hepatitis C virus core protein promotes immortalization of primary human hepatocytes. Virology. 2000;271:197-204. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 126] [Cited by in F6Publishing: 136] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 166. | Lim JS, Park SH, Jang KL. Hepatitis C virus Core protein overcomes stress-induced premature senescence by down-regulating p16 expression via DNA methylation. Cancer Lett. 2012;321:154-161. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 44] [Cited by in F6Publishing: 45] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 167. | Hassan M, Selimovic D, Ghozlan H, Abdel-kader O. Hepatitis C virus core protein triggers hepatic angiogenesis by a mechanism including multiple pathways. Hepatology. 2009;49:1469-1482. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 81] [Cited by in F6Publishing: 83] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 168. | Battaglia S, Benzoubir N, Nobilet S, Charneau P, Samuel D, Zignego AL, Atfi A, Bréchot C, Bourgeade MF. Liver cancer-derived hepatitis C virus core proteins shift TGF-beta responses from tumor suppression to epithelial-mesenchymal transition. PLoS One. 2009;4:e4355. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 88] [Cited by in F6Publishing: 99] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 169. | Quan H, Zhou F, Nie D, Chen Q, Cai X, Shan X, Zhou Z, Chen K, Huang A, Li S. Hepatitis C virus core protein epigenetically silences SFRP1 and enhances HCC aggressiveness by inducing epithelial-mesenchymal transition. Oncogene. 2014;33:2826-2835. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 74] [Cited by in F6Publishing: 83] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 170. | Feng X, Xiu B, Xu L, Yang X, He J, Leong D, He F, Zhang H. Hepatitis C virus core protein promotes the migration and invasion of hepatocyte via activating transcription of extracellular matrix metalloproteinase inducer. Virus Res. 2011;158:146-153. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 10] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 171. | Jahan S, Khaliq S, Ijaz B, Ahmad W, Hassan S. Role of HCV Core gene of genotype 1a and 3a and host gene Cox-2 in HCV-induced pathogenesis. Virol J. 2011;8:155. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 27] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 172. | Rüster B, Zeuzem S, Krump-Konvalinkova V, Berg T, Jonas S, Severin K, Roth WK. Comparative sequence analysis of the core- and NS5-region of hepatitis C virus from tumor and adjacent non-tumor tissue. J Med Virol. 2001;63:128-134. [PubMed] [Cited in This Article: ] |
| 173. | Alam SS, Nakamura T, Naganuma A, Nozaki A, Nouso K, Shimomura H, Kato N. Hepatitis C virus quasispecies in cancerous and noncancerous hepatic lesions: the core protein-encoding region. Acta Med Okayama. 2002;56:141-147. [PubMed] [Cited in This Article: ] |
| 174. | Ogata S, Nagano-Fujii M, Ku Y, Yoon S, Hotta H. Comparative sequence analysis of the core protein and its frameshift product, the F protein, of hepatitis C virus subtype 1b strains obtained from patients with and without hepatocellular carcinoma. J Clin Microbiol. 2002;40:3625-3630. [PubMed] [Cited in This Article: ] |
| 175. | Fishman SL, Factor SH, Balestrieri C, Fan X, Dibisceglie AM, Desai SM, Benson G, Branch AD. Mutations in the hepatitis C virus core gene are associated with advanced liver disease and hepatocellular carcinoma. Clin Cancer Res. 2009;15:3205-3213. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 57] [Cited by in F6Publishing: 62] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 176. | Fukuhara T, Takeishi K, Toshima T, Morita K, Ueda S, Iguchi T, Nagata S, Sugimachi K, Ikegami T, Gion T. Impact of amino acid substitutions in the core region of HCV on multistep hepatocarcinogenesis. Hepatol Res. 2010;40:171-178. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 9] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 177. | Hu Z, Muroyama R, Kowatari N, Chang J, Omata M, Kato N. Characteristic mutations in hepatitis C virus core gene related to the occurrence of hepatocellular carcinoma. Cancer Sci. 2009;100:2465-2468. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in F6Publishing: 30] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 178. | Kobayashi M, Akuta N, Suzuki F, Hosaka T, Sezaki H, Kobayashi M, Suzuki Y, Arase Y, Ikeda K, Watahiki S. Influence of amino-acid polymorphism in the core protein on progression of liver disease in patients infected with hepatitis C virus genotype 1b. J Med Virol. 2010;82:41-48. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 25] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 179. | Guo N, Chen R, Li Z, Liu Y, Cheng D, Zhou Q, Zhou J, Lin Q. Hepatitis C virus core upregulates the methylation status of the RASSF1A promoter through regulation of SMYD3 in hilar cholangiocarcinoma cells. Acta Biochim Biophys Sin (Shanghai). 2011;43:354-361. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 25] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 180. | García-Samaniego J, Rodríguez M, Berenguer J, Rodríguez-Rosado R, Carbó J, Asensi V, Soriano V. Hepatocellular carcinoma in HIV-infected patients with chronic hepatitis C. Am J Gastroenterol. 2001;96:179-183. [PubMed] [Cited in This Article: ] |
| 181. | Khan KA, Abbas W, Varin A, Kumar A, Di Martino V, Dichamp I, Herbein G. HIV-1 Nef interacts with HCV Core, recruits TRAF2, TRAF5 and TRAF6, and stimulates HIV-1 replication in macrophages. J Innate Immun. 2013;5:639-656. [PubMed] [Cited in This Article: ] |
| 182. | Vivithanaporn P, Maingat F, Lin LT, Na H, Richardson CD, Agrawal B, Cohen EA, Jhamandas JH, Power C. Hepatitis C virus core protein induces neuroimmune activation and potentiates Human Immunodeficiency Virus-1 neurotoxicity. PLoS One. 2010;5:e12856. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 60] [Cited by in F6Publishing: 64] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 183. | Donato F, Boffetta P, Puoti M. A meta-analysis of epidemiological studies on the combined effect of hepatitis B and C virus infections in causing hepatocellular carcinoma. Int J Cancer. 1998;75:347-354. [PubMed] [Cited in This Article: ] |
| 184. | Liu W, Chen JR, Hsu CH, Li YH, Chen YM, Lin CY, Huang SJ, Chang ZK, Chen YC, Lin CH. A zebrafish model of intrahepatic cholangiocarcinoma by dual expression of hepatitis B virus X and hepatitis C virus core protein in liver. Hepatology. 2012;56:2268-2276. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 52] [Cited by in F6Publishing: 56] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 185. | Martoglio B, Dobberstein B. Signal sequences: more than just greasy peptides. Trends Cell Biol. 1998;8:410-415. [PubMed] [Cited in This Article: ] |
| 186. | Martoglio B. Intramembrane proteolysis and post-targeting functions of signal peptides. Biochem Soc Trans. 2003;31:1243-1247. [PubMed] [Cited in This Article: ] |
| 187. | Hombach J, Pircher H, Tonegawa S, Zinkernagel RM. Strictly transporter of antigen presentation (TAP)-dependent presentation of an immunodominant cytotoxic T lymphocyte epitope in the signal sequence of a virus protein. J Exp Med. 1995;182:1615-1619. [PubMed] [Cited in This Article: ] |
| 188. | Kammer AR, van der Burg SH, Grabscheid B, Hunziker IP, Kwappenberg KM, Reichen J, Melief CJ, Cerny A. Molecular mimicry of human cytochrome P450 by hepatitis C virus at the level of cytotoxic T cell recognition. J Exp Med. 1999;190:169-176. [PubMed] [Cited in This Article: ] |
| 189. | Gao DY, Jin GD, Yao BL, Zhang DH, Gu LL, Lu ZM, Gong Q, Lone YC, Deng Q, Zhang XX. Characterization of the specific CD4+ T cell response against the F protein during chronic hepatitis C virus infection. PLoS One. 2010;5:e14237. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 8] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 190. | Vassilaki N, Friebe P, Meuleman P, Kallis S, Kaul A, Paranhos-Baccalà G, Leroux-Roels G, Mavromara P, Bartenschlager R. Role of the hepatitis C virus core+1 open reading frame and core cis-acting RNA elements in viral RNA translation and replication. J Virol. 2008;82:11503-11515. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 79] [Cited by in F6Publishing: 79] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 191. | Ray RB, Steele R, Basu A, Meyer K, Majumder M, Ghosh AK, Ray R. Distinct functional role of Hepatitis C virus core protein on NF-kappaB regulation is linked to genomic variation. Virus Res. 2002;87:21-29. [PubMed] [Cited in This Article: ] |
| 192. | Basu A, Steele R, Ray R, Ray RB. Functional properties of a 16 kDa protein translated from an alternative open reading frame of the core-encoding genomic region of hepatitis C virus. J Gen Virol. 2004;85:2299-2306. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 28] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 193. | Ma HC, Lin TW, Li H, Iguchi-Ariga SM, Ariga H, Chuang YL, Ou JH, Lo SY. Hepatitis C virus ARFP/F protein interacts with cellular MM-1 protein and enhances the gene trans-activation activity of c-Myc. J Biomed Sci. 2008;15:417-425. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 31] [Cited by in F6Publishing: 32] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 194. | Yeh CT, Lo SY, Dai DI, Tang JH, Chu CM, Liaw YF. Amino acid substitutions in codons 9-11 of hepatitis C virus core protein lead to the synthesis of a short core protein product. J Gastroenterol Hepatol. 2000;15:182-191. [PubMed] [Cited in This Article: ] |
| 195. | Dalagiorgou G, Vassilaki N, Foka P, Boumlic A, Kakkanas A, Kochlios E, Khalili S, Aslanoglou E, Veletza S, Orfanoudakis G. High levels of HCV core+1 antibodies in HCV patients with hepatocellular carcinoma. J Gen Virol. 2011;92:1343-1351. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 25] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 196. | Kotta-Loizou I, Vassilaki N, Pissas G, Kakkanas A, Bakiri L, Bartenschlager R, Mavromara P. Hepatitis C virus core+1/ARF protein decreases hepcidin transcription through an AP1 binding site. J Gen Virol. 2013;94:1528-1534. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 16] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 197. | Budkowska A, Kakkanas A, Nerrienet E, Kalinina O, Maillard P, Horm SV, Dalagiorgou G, Vassilaki N, Georgopoulou U, Martinot M. Synonymous mutations in the core gene are linked to unusual serological profile in hepatitis C virus infection. PLoS One. 2011;6:e15871. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 16] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 198. | Tsao ML, Chao CH, Yeh CT. Interaction of hepatitis C virus F protein with prefoldin 2 perturbs tubulin cytoskeleton organization. Biochem Biophys Res Commun. 2006;348:271-277. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in F6Publishing: 30] [Article Influence: 1.7] [Reference Citation Analysis (0)] |