Copyright
©2014 Baishideng Publishing Group Inc.
World J Gastroenterol. May 21, 2014; 20(19): 5773-5793
Published online May 21, 2014. doi: 10.3748/wjg.v20.i19.5773
Published online May 21, 2014. doi: 10.3748/wjg.v20.i19.5773
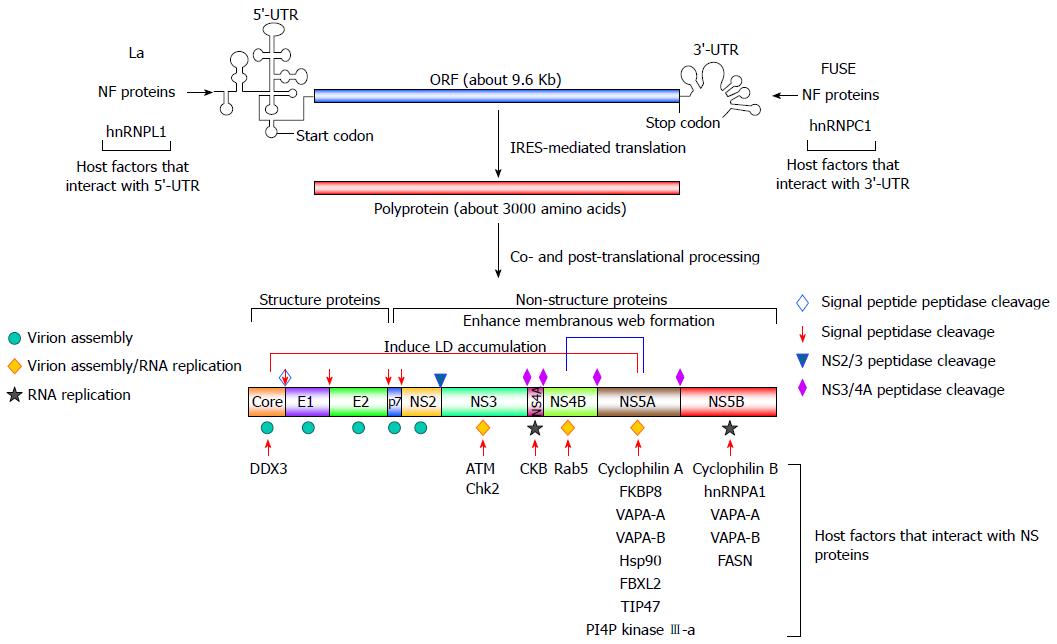
Figure 2 Genome organization of hepatitis C virus and cellular proteins that modulate viral RNA replication.
The positive-sense, single-stranded hepatitis C virus (HCV) genome, containing an open reading frame (ORF) of 9.6 Kb that is flanked by the 5’- and 3’ untranslated regions (UTR), is translated using an internal ribosome entry site (IRES) to a polyprotein, which is processed by cellular and viral proteases to mature into three structural proteins (core, envelope glycoproteins E1 and E2) and seven nonstructural proteins (p7, NS2, NS3, NS4A, NS4B, NS5A, and NS5B). The role of each viral protein in virus assembly, RNA replication, or both is indicated by the symbol shown on the left. The cellular proteins that modulate HCV replication and their interacting viral counterparts are shown. Host factors that interact with the 5’-UTR and 3’-UTR, as indicated, regulate IRES-mediated translation and viral RNA replication, respectively. Membrane-associated proteins, as shown, modulate HCV replication through interactions with NS5A and NS5B. Other cellular factors, such as DDX3, ATM and Chk2, CKB, and Rab5, promote the replication of HCV viral RNA via interacting with core, NS3, NS4A, and NS4B, respectively.
- Citation: Ke PY, Chen SSL. Autophagy in hepatitis C virus-host interactions: Potential roles and therapeutic targets for liver-associated diseases. World J Gastroenterol 2014; 20(19): 5773-5793
- URL: https://www.wjgnet.com/1007-9327/full/v20/i19/5773.htm
- DOI: https://dx.doi.org/10.3748/wjg.v20.i19.5773