Copyright
©2014 Baishideng Publishing Group Co.
World J Gastroenterol. Apr 14, 2014; 20(14): 3804-3824
Published online Apr 14, 2014. doi: 10.3748/wjg.v20.i14.3804
Published online Apr 14, 2014. doi: 10.3748/wjg.v20.i14.3804
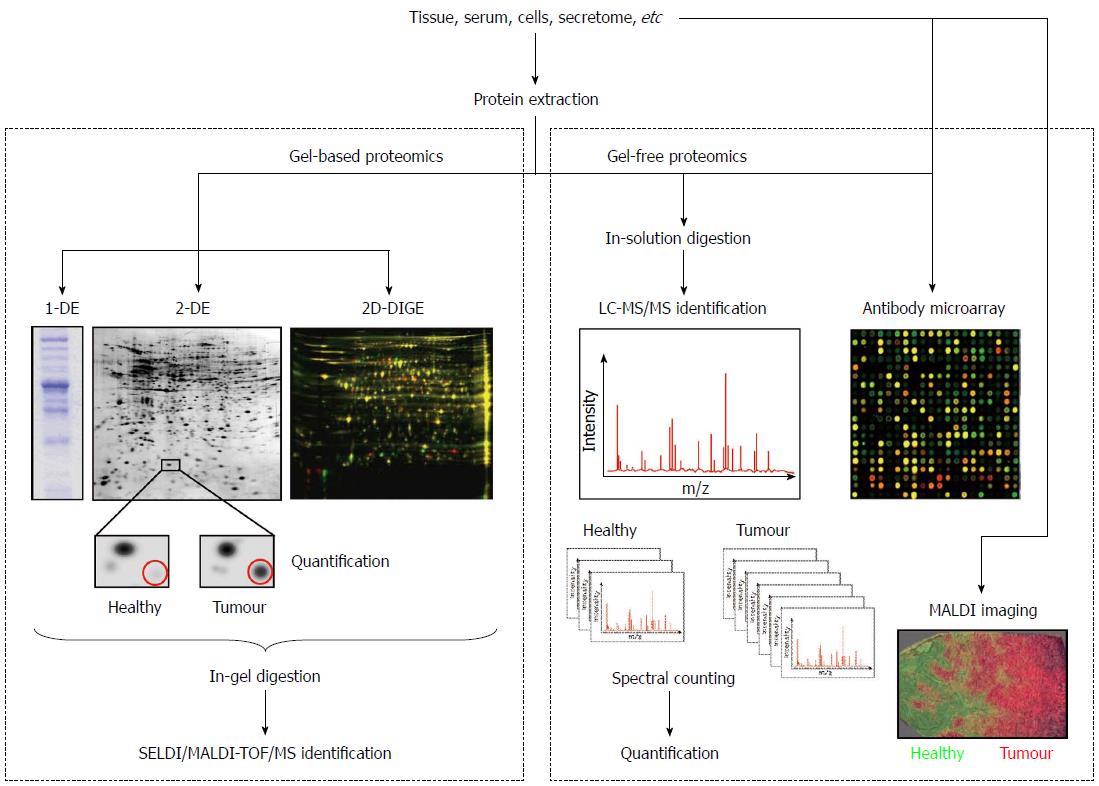
Figure 1 Schematic representation of the principal workflows used in proteomics for colorectal cancer biomarker discovery.
Please note that for both gel-based and gel-free proteomics only one of the quantification methods is shown. DE: Dimensional electrophoresis; 2D-DIGE: Two-dimensional differential in-gel electrophoresis; SELDI: Surface-enhanced laser desorption/ionization; MALDI: Matrix-assisted laser desorption/ionization; TOF: Time of flight; MS: Mass spectrometry.
- Citation: Álvarez-Chaver P, Otero-Estévez O, Páez de la Cadena M, Rodríguez-Berrocal FJ, Martínez-Zorzano VS. Proteomics for discovery of candidate colorectal cancer biomarkers. World J Gastroenterol 2014; 20(14): 3804-3824
- URL: https://www.wjgnet.com/1007-9327/full/v20/i14/3804.htm
- DOI: https://dx.doi.org/10.3748/wjg.v20.i14.3804