Copyright
©2014 Baishideng Publishing Group Co.
World J Gastroenterol. Apr 7, 2014; 20(13): 3401-3409
Published online Apr 7, 2014. doi: 10.3748/wjg.v20.i13.3401
Published online Apr 7, 2014. doi: 10.3748/wjg.v20.i13.3401
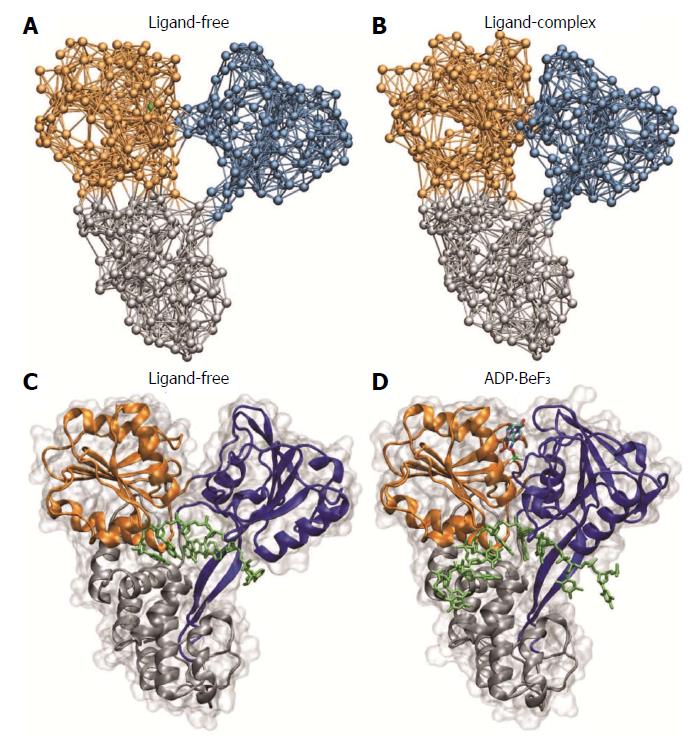
Figure 5 Comparison with novel hepatitis C virus crystal structures.
Structural changes between ligand-free conformation and ligand-bound complex as predicted by our simulations (A and B) are compared with structures determined in recent experiments (C and D, Protein Data Bank codes 3KQH and 3KQU; ADP.BeF3 was used as ATP analog). ATP: Adenosine-triphosphate; ADP: Adenosine-diphosphate.
- Citation: Flechsig H. Computational biology approach to uncover hepatitis C virus helicase operation. World J Gastroenterol 2014; 20(13): 3401-3409
- URL: https://www.wjgnet.com/1007-9327/full/v20/i13/3401.htm
- DOI: https://dx.doi.org/10.3748/wjg.v20.i13.3401