Published online Apr 7, 2014. doi: 10.3748/wjg.v20.i13.3457
Revised: December 15, 2013
Accepted: January 3, 2014
Published online: April 7, 2014
Processing time: 187 Days and 17.4 Hours
Hepatitis C virus (HCV) infection is a global health problem, with an estimated 170 million people being chronically infected. HCV cell entry is a complex multi-step process, involving several cellular factors that trigger virus uptake into the hepatocytes. The high- density lipoprotein receptor scavenger receptor class B type I, tetraspanin CD81, tight junction protein claudin-1, and occludin are the main receptors that mediate the initial step of HCV infection. In addition, the virus uses cell receptor tyrosine kinases as entry regulators, such as epidermal growth factor receptor and ephrin receptor A2. This review summarizes the current understanding about how cell surface molecules are involved in HCV attachment, internalization, and membrane fusion, and how host cell kinases regulate virus entry. The advances of the potential antiviral agents targeting this process are introduced.
Core tip: Cell entry is the first step in viral infection and replication, which offers an important target for antiviral therapy. Hepatitis C virus (HCV) cell entry is a complex multi-step process, involving a several cellular factors that trigger virus uptake into the hepatocytes. This review summarizes the current understanding about how cell surface molecules are involved in HCV attachment, internalization, and membrane fusion, and how host cell kinases regulate virus entry. The advances of the potential antiviral agents targeting this process are introduced.
- Citation: Zhu YZ, Qian XJ, Zhao P, Qi ZT. How hepatitis C virus invades hepatocytes: The mystery of viral entry. World J Gastroenterol 2014; 20(13): 3457-3467
- URL: https://www.wjgnet.com/1007-9327/full/v20/i13/3457.htm
- DOI: https://dx.doi.org/10.3748/wjg.v20.i13.3457
Hepatitis C virus (HCV) infection is a global health problem. About 130-170 million people are chronically infected and at risk of developing liver cirrhosis and/or liver cancer[1]. More than 350000 people die from hepatitis-C-related liver diseases every year. Current antiviral treatment of chronic HCV infection is based on pegylated interferon (PEG-IFN) and ribavirin, which is limited by side effects and suboptimal response rates[2]. Since 2011, standard of care (SOC) treatment of HCV includes the addition of direct-acting antivirals with a protease inhibitor to the PEG-IFN and ribavirin[3,4]. However, resistance and severe side effects are still issues[5]. There is currently no vaccine for hepatitis C. Thus, novel preventive and therapeutic strategies are urgently needed.
Cell entry is the first step in viral infection and replication, which offers an important target for antiviral therapy. The establishment of infectious HCV pseudoparticles (HCVpps)[6,7], cell-culture-derived HCV (HCVcc)[8-10], and progress in small animal models of HCV infection[11-14] have allowed us to identify host factors contributing to HCV entry and to develop antiviral compounds.
HCV entry into hepatocytes is thought to be a highly coordinated process that involves several cell surface molecules in sequential steps[15]. More than two decades of intense research has provided a detailed understanding of these entry factors, and considerable progress has been made to decipher how they trigger and facilitate HCV cell entry. Recent data also demonstrate the important role of host cell tyrosine protein kinases in regulating HCV entry[16]. In this review, we summarize the current knowledge about how HCV uses these host factors to invade hepatocytes, and how host cell kinases regulate virus entry, and the recent progress of antiviral therapy targeting this process.
HCV is an enveloped, positive-stranded RNA virus belonging to the Hepacivirus in the Flaviviridae family. The HCV virion is comprised of a nucleocapsid surrounded by a host-derived membrane containing the E1 and E2 HCV glycoproteins[17]. Functional virion-associated E1 and E2 are thought to form a non-covalent heterodimer stabilized by disulfide bridges[18,19], which mediate the majority of cell-entry processes of the virion, including interaction with host receptors and fusion in the low pH environment of early endosomes[20-23]. The first 27 amino acids residues of the N terminus of E2, called hypervariable region 1 (HVR1), are the most divergent among HCV isolates. Recent data show that HVR1 contains three different functional microdomains that cooperate to confer HCV cell entry and immune evasion[24]. Based on similarities with envelope proteins of the Flaviviridae family, E2 was previously proposed to be the fusion protein[25-27]. However, recent data on pestiviruses and HCV are no longer in favor of E2 being a fusion protein. The structure of E2 protein from the pestivirus bovine viral diarrhea virus does not show any evidence of a fusion peptide in this protein[28,29]. More recently, the crystal structure of the E2 core bound to broadly neutralizing antibody AR3C was resolved and revealed a compact architecture composed of a central immunoglobulin-fold β sandwich flanked by two additional protein layers, which differs markedly from predictions of an extended, three-domain, class II fusion protein fold[25]. E1 was recently demonstrated to be a modulator of HCV binding to receptors and membrane fusion[30]. The characterization of HCV particles circulating in patient sera is the heterogeneity of size and density, due to the association of virions with different serum components, such as immunoglobulins[31] and different classes of lipoproteins[32].The nature of the association between HCV and lipoproteins remains unclear. Circulating virions might directly bind to low-density lipoprotein (LDL) and very low-density lipoprotein (VLDL) in the blood, or interact with lipoprotein components during virus assembly and are secreted together with VLDL[33]. Highly infectious HCV particles are associated with VLDL, termed lipoviral particles (LVPs). LVPs are lipoprotein-like structures composed of triglyceride-rich lipoproteins containing apolipoprotein (Apo)B, ApoE and ApoC1, viral nucleocapsids, and envelope glycoproteins E1 and E2[34-38]. Several functional roles for the formation of HCV-lipoprotein complexes have been proposed, including interacting with lipoprotein receptors for attachment and entry, concealing epitopes to facilitate immune escape, and hijacking host factors for HCV maturation and secretion[39].
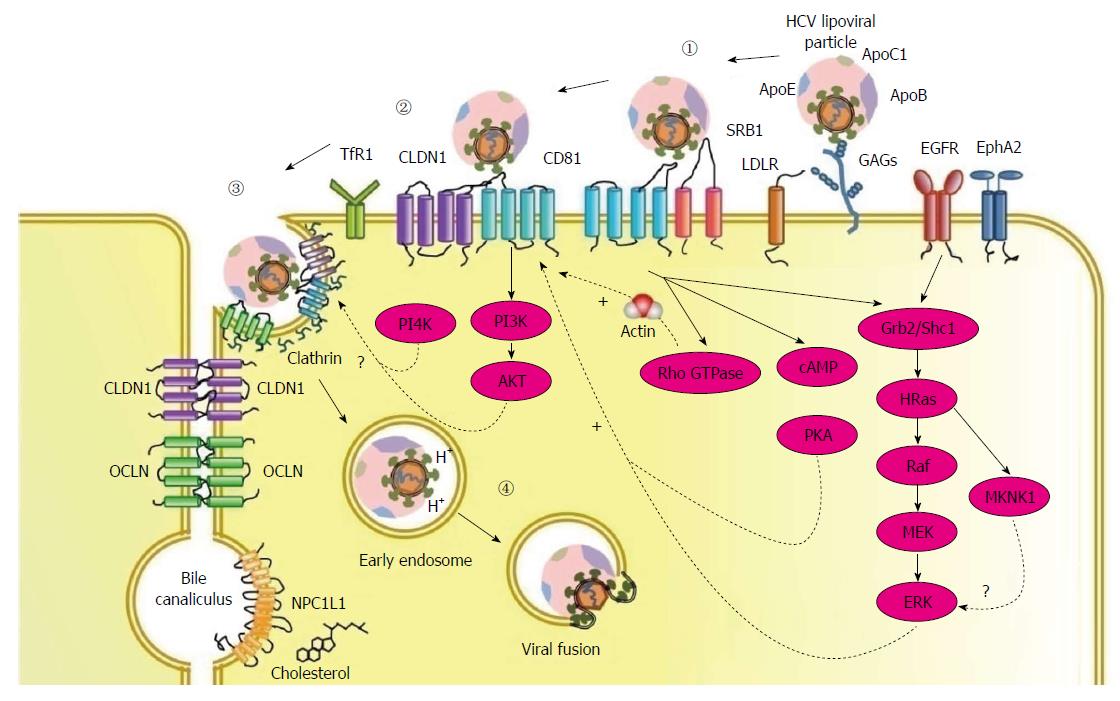
The HCV entry process into human hepatocytes requires numerous host proteins, including two binding factors glycosaminoglycans (GAGs)[40,41] and the LDL receptor (LDLR)[42-45], four receptors scavenger receptor class B type I (SR-BI)[46], tetraspanin CD81[47], tight junction proteins, claudin-1 (CLDN1)[48] and occludin (OCLN)[49], and several entry factors including epidermal growth factor receptor (EGFR), ephrin receptor A2 (EphA2)[50], transferrin receptor 1 (TfR1)[51], and cholesterol transporter Niemann-Pick C1-like 1 (NPC1L1)[52] (Table 1). The entry pathway consists of three steps: (1) viral attachment to the hepatocyte; (2) receptor-mediated endocytosis of the viral particle; and (3) endosomal fusion (Figure 1).
| Characterization | Main function in HCV entry | |
| GAGs | Binding factor | Attachment to HCV E1 and ApoE |
| LDL-R | Binding factor | Binding to virion-associated lipoprotein |
| SR-BI | Receptor | Binding to virion-associated lipoprotein; lipid transfer; binding to E2 HVR1 |
| CD81 | Receptor | Interaction with E2; signaling pathway activation which promotes actin remodeling |
| CLDN1 | Receptor | Interaction with HCV-CD81 complex |
| OCLN | Receptor | Function at the late stage of HCV entry |
| EGFR and EphA2 | Entry factor | Signaling pathways activation which lead to the formation of CD81-CLDN1 complexes |
| TfR1 | Entry factor | Possibly involved in virus internalization |
| NPC1L1 | Entry factor | Possibly involved in endosomal fusion |
In vivo, circulating HCV enters the liver through the sinusoidal blood. The liver sinusoidal endothelial cells (LSECs) and Kupffer cells express liver/lymph-node-specific intercellular adhesion molecule 3-grabbing integrin (L-SIGN) and dendritic-cell-specific intercellular adhesion molecule-3-grabbing non-integrin (DC-SIGN), respectively. Both L-SIGN and DC-SIGN bind HCV E2 with high affinity, suggesting they may capture particles from blood and transmit virus to hepatocytes[53,54].
Successful traverse from the endothelium allows the virus to contact hepatocytes at the basal surface. GAGs and LDLR are proposed to contribute to concentrate HCV particles on target cell surface[41,44]. On hepatocytes, highly sulfated GAGs (HS GAGs) serve as the first attachment sites[55]. While direct E2 binding to HS GAGs is observed[55], E1 and ApoE are also suggested to attach to GAGs[56].
As a result of the association between HCV and lipoproteins, the LDLR has been proposed as another potential attachment factor for HCV[43]. Although HCV particles bind to the LDLR, it seems that this interaction does not lead to a productive entry process[57].
After initial attachment to the host cell surface, the virus interacts with co-receptors and other specific entry factors, which leads to molecular rearrangements at the plasma membrane and subsequently results in viral internalization.
SR-BI is highly expressed in the liver and steroidogenic tissues[58], can bind a variety of lipoproteins including high-density lipoprotein (HDL), LDL, and oxidized LDL[59], and plays a key role in mediating selective cholesterol uptake from HDL into hepatocytes to maintain lipid homeostasis[60]. SR-BI is a glycoprotein with two N- and C-terminal cytoplasmic domains separated by a large extracellular domain that is involved in lipid metabolism. The large extracellular loop of SR-BI was initially identified to bind directly to the HVR1 of HCV E2[61], and then interact with virus-associated lipoproteins[62]. Recent data show that HCV particles utilize SR-BI in a manifold manner[63]. First, the initial attachment of HCV particles to SR-BI is independent of E2 but, rather, is mediated by lipoprotein components, such as ApoE[63,64]. Second, the lipid transfer function of SR-BI may facilitate the viral particles access to the next entry step, which would allow exposure of CD81 binding sites on HCV E2 and transfer the virus particle to CD81[63]. Finally, direct interactions between E2 HVR1 and SR-BI enhance infectivity of the particle at post-attachment levels[65].
CD81 is a member of the tetraspanin superfamily defined by four transmembrane domains, a conserved CCG motif, and four cysteine residues that form critical disulfide bonds in the large extracellular loop (LEL). CD81 interacts with HCV E2 glycoprotein via a series of discontinuous amino acid residues in the LEL[66]. Several studies suggest that CD81 acts as a post-binding entry molecule. HCV-CD81 interaction induces a conformational rearrangement of the E1 and E2 glycoproteins that facilitates pH-dependent fusion and endocytosis of the virus[67]. Upon CD81 engagement, HCV activates signals allowing the lateral movement of the virus particle/CD81 complex and its delivery to areas of cell-cell contact[68]. Tetraspanins associate with each other to carry out many biological functions, and several CD81 partners and CD81-associated tetraspanins are proposed to play a role in HCV entry[69-71].
CLDN1 is a member of the claudin family of tight junction proteins, consisting of a large and small extracellular loop (EL1 and EL2, respectively), and four transmembrane domains. The highly conserved EL1 is critical for HCV entry[48]. CLDN6 and CLDN9 can replace CLDN1 for HCV entry, but they are expressed only at low levels in the liver[72].
CD81 and CLDN1 act at closely related time-points during HCV entry[73], and direct CD81-CLDN1 interaction is observed[74], strongly suggesting they form a complex to facilitate virus entry. The HCV envelope glycoproteins do not directly interact with CLDN1, but CLDN1 interacts with CD81 and thereby plays an important role during post-binding steps of the HCV entry process[48,73,74]. CLDN1 is expressed both at the basolateral and apical membranes[75]. Interestingly, CLDN1-CD81 complexes are absent from apically located tight junctions, suggesting that virus engagement of basolateral pools of CD81 and CLDN1 may initiate the particle internalization process[75].
EGFR is a receptor tyrosine kinase (RTK) that regulates several key processes, including cell proliferation, survival, and differentiation during development, tissue homeostasis and tumorigenesis[76]. EphA2 mediates cell positioning, cell morphology, polarity and motility[77]. EGFR and EphA2 were recently identified to be required for HCV entry[50] and modulate interactions between CD81 and CLDN1 by EGFR-dependent signaling pathways[78].
OCLN, also belongs to the tight junction protein family, and has been identified as another co-receptor required for a late post-binding event[49]. OCLN is composed of four transmembrane regions, two extracellular loops and N- and C-terminal cytoplasmic regions[79]. The determinants on OCLN of HCV infectivity and species specificity are mapped to the second extracellular loop[80]. Although OCLN has been reported to interact with the HCV E2 glycoproteins[81], whether the interaction is mediated via a direct E2-OCLN binding or by indirect interactions with CD81/CLDN1 complexes is unclear.
Internalization of HCV particles is dependent on clathrin-mediated endocytosis[22]. HCV promotes CD81 endocytosis via a clathrin- and dynamin-dependent process in association with CLDN1[82].
Recently, TfR1 was identified as a HCV entry factor[51]. TfR1 is ubiquitously expressed in all tissues and is the main receptor for cellular iron uptake into cells via clathrin-mediated endocytosis. TfR1 plays a role in HCV infection at the level of glycoprotein-mediated entry, acts after CD81, and possibly is involved in HCV particle internalization[51].
The fusion within the endosomal cell compartment between viral and host membranes is the final step of HCV entry to release the viral RNA for the following HCV life cycle. It is believed that this process is triggered in a receptor-independent but pH-dependent manner, and is closely linked with lipid composition[83]. The virus-receptor interactions and low pH might cause glycoprotein rearrangements to trigger fusion-related protein transition from a pre-fusion state to a post-fusion structure[84].
Of the two envelope protein, E1 acts as a protein chaperone. It has been shown that specific residues of fusion-peptide-like domain on E1 are required for mediating cell fusion and entry of HCV[27,85,86]. Still, the role of this domain in HCV entry needs further investigation. However, the role of E2 in viral fusion is controversial. Previous studies have suggested that a specific region of E2 is a key fusion determinant of HCV[26,27], while Law’s team has found no presence of a fusion peptide in HCV E2 by crystal structure analysis[25]. The conformational changes of the envelope proteins finally lead to a fusion pore to release the nucleocapsid into the cytosol[87,88].
Besides the acidic endosomal pH environment, the cholesterol of the target membranes is also found to have a strong promoting effect on the membrane fusion capacity of flaviviruses[89-91]. The NPC1L1 cholesterol uptake receptor has been identified recently to be one of key entry factors for HCV[52]. It has been revealed that it might promote HCV entry by modulating cholesterol homeostasis or influencing cholesterol level of LVPs, thus affecting membrane composition required for membrane fusion[92].
It has been recently demonstrated that HCV uses host kinases during the entry process to facilitate virus-receptor interaction, and appropriate membrane traffic.
There is strong evidence that CD81 and CLDN-1 work together to facilitate HCV entry. The association of CD81 and CLDN-1 appears to be regulated by multiple signaling pathways. Using a functional RNAi kinase screen, two RTKs, EGFR and EphA2, are identified as host cofactors for HCV entry by prompting CD81-CLDN1 complex formation[50]. EGFR regulates CD81-CLDN1 co-receptor association by the activation of the EGFR/Shc1/Grb2/HRas signaling pathway[78]. Proteomic analysis has revealed that HRas associates with CD81 and CLDN1, suggesting HRas acts as a molecular switch promoting RTK-mediated HCV entry[78]. Following EGFR stimulation, the Ras/MEK/ERK pathway is activated, which could lead to the activation of MAPK interacting serine/threonine kinase 1 (MKNK1)[93]. MKNK1 was recently identified as a host factor in HCV entry, which possibly acts to facilitate the downstream effect of EGFR[93].
For a multi-receptor virus, successful entry is based on the lateral movement of the virus on the plasma membrane and interaction with sufficient receptors to initiate internalization[94]. In the case of HCV, after the interaction with HCV E2, CD81 activates Rho GTPase family members Rac, Rho and cell division cycle 42 that promote actin remodeling, thus allowing the delivery of the E2-CD81 complex to come into contact with CLDN1[68].
If the localization of either co-receptor is disrupted, the association of CD81 with CLDN1 will be perturbed[95]. By screening a series of kinase inhibitors for their effects on HCV infection, protein kinase A (PKA) was identified as having an important role in HCV entry[96]. Initiation of HCV infection activates PKA in a cAMP-dependent manner to retain CLDN1 on the plasma membrane, which then promotes the entry of HCV[96].
The class I phosphatidylinositol 3-kinase (PI3K) is activated by G-protein-coupled receptors and tyrosine kinase receptors. Upon its activation, it converts phosphatidylinositol 4,5-biphosphate to phosphatidylinositol 3,4,5-triphosphate, which binds to and recruits AKT to the membrane for its phosphorylation. Many flaviviruses regulate the PI3K-AKT pathway for their entry[97,98]. HCV rapidly and transiently activates the PI3K-AKT pathway in the early stage of infection to enhance its entry, and the activation is mediated by the interaction between HCV E2 and its co-receptors, CD81 and CLDN1[99]. However, the exact step of the entry process that the AKT pathway regulates remains elusive.
In addition, phosphatidylinositol 4-kinases type III α (PI4KIIIα) and β (PI4KIIIβ) have been suggested to play a role in membrane remodeling and trafficking during HCV entry[100,101]. However, the underlying molecular mechanism is unclear.
HCV entry has been revealed as a highly complex process requiring the involvement of viral envelope proteins and multiple host proteins, which offer a large number of promising therapeutic targets (Table 2). Each stage of HCV entry can be designed as an antiviral target, including virus particles, virus attachment, receptor-mediated endocytosis, endosomal fusion, and the regulatory pathways.
| Stages of HCV entry process | Target | Antiviral agents | Ref. |
| HCV particle/attachment | HCV E1 and E2 | Neutralizing antibodies | [102,103] |
| Heparin | [41,55] | ||
| Lectins | [106,107] | ||
| EGCG | [109,110] | ||
| Oleanane-type triterpenes | [111] | ||
| Virion-associated lipoprotein | Anti-apoE mAb | [108] | |
| Receptor-mediated endocytosis | SRBI | Anti-SR-BI mAb | [113] |
| ITX 5061 | [117] | ||
| CD81 | Anti-CD81 mAb | [114] | |
| CLDN1 | Anti-CLDN1mAb | [115] | |
| CLDN1-derived peptide | [116] | ||
| EGFR | Erlotinib | [50] | |
| NPC1L1 | Ezetimibe | [52] | |
| TfR1 | Anti-TfR1 mAb | [51] | |
| Internalization | Amphipathic DNA polymers | [118] | |
| Arbidol | [122] | ||
| Tamoxifen | [123] | ||
| Endosomal fusion | Fusion | E2-derived peptide | [121] |
| CD81-derived peptide | [124] | ||
| Curcumin | [125] | ||
| Phenothiazines | [126] | ||
| Ferroquine | [127] | ||
| aUY11 | [128] | ||
| HCV II-1/GS-563253 | [129] | ||
| Regulation | Ras | Tipifarnib | [78] |
| PKA | H89 | [96] | |
| MKNK1 | RO4475417 | [93] | |
| PI3K | Wortmanin | [99] |
HCV entry can be inhibited with monoclonal or polyclonal neutralizing antibodies[102-104]. However, cross-reactive neutralizing antibodies seem to appear later during HCV infection, and several mechanisms contribute to reduce their accessibility to their cognate epitopes. These include the masking of major conserved neutralizing epitopes by HVR1, specific N-linked glycans, and the lipid moiety of the viral particle. Other potential mechanisms of evasion from the neutralizing antibody response include a modulation by HDLs and interfering antibodies, as well as the capacity of the virus to be transferred by cell-to-cell contact[105].
Several non-HCV specific molecules interfering with HCV envelope glycoproteins and abrogating viral attachment have been described. Heparin, a structural analog of GAGs, has been demonstrated to inhibit HCV entry[41,55]. Lectins, such as cyanovirin-N and griffithsin, bind to the high-mannose glycans present on the HCV particles and thereby inhibit HCV entry[106,107]. ApoE antibodies and peptides are able to inhibit HCV entry by blocking its binding to target cells[108]. Some natural products have been shown to prevent virus binding, such as the green tea polyphenol epigallocatechin-3-gallate[109,110] and oleanane-type trierpene[111].
To overcome the high variability of the viral envelope proteins, small-molecule host-targeting agents (HTAs) are also being investigated[112]. Antibodies or peptides targeting CD81, SR-B1 and CLDN1 have been shown to be an efficacious way to prevent HCV infection both in vitro and in vivo in a genotype-independent manner[113-116]. ITX 5061, one of the most promising HTAs, which promotes HDL levels in animals and patients by targeting SRB1[117], is currently in phase 2 clinical trials. Clathrin-dependent endocytosis[118-120] and endosomal fusion[121] are also potential targets for the development of anti-HCV compounds. Arbidol suppresses clathrin-mediated endocytosis by hindering HCV endosomal trafficking and impairing dynamin-2-induced membrane scission[122]. Tamoxifen is a selective estrogen receptor modulator, and affects both viral binding and post-binding events including endocytosis[123]. The stapled peptides are designed according to the linear peptide sequence of the large extracellular loop of CD81, which has been reported to play an important role in HCV E2 binding interaction, and has an inhibitory effect on HCV membrane fusion[124]. Curcumin affects membrane fluidity, therefore impeding viral binding and fusion[125]. Phenothiazines inhibit HCV infection at an early stage of the viral cycle by interfering with virion-cell fusion[126]. They insert into cholesterol-rich domains of target membranes and perturb cholesterol distribution in lipid membranes, creating a barrier to virion-cell fusion. Ferroquine, an anolog of chloroquine, suppresses HCV infection at a late post-binding step by impairing the fusion process[127]. An arabino-based rigid amphipathic fusion inhibitor, aUY11, inhibits HCV infection at the early fusion process by interacting with envelope lipids[128]. HCV-II/GS-563253 suppresses HCV endosomal fusion by locking the viral envelope pre-fusion state or formation of an incapable envelope conformation for viral fusion[129].
Given the relevance of host cell kinases for HCV entry and the number of kinase inhibitors being developed to treat a wide variety of human diseases, kinase inhibitors have been suggested as a novel class of antivirals for the prevention and treatment of HCV infection[78,93,96,99]. Erlotinib, a clinically approved inhibitor of EGFR, and the NPC1L1 inhibitor ezetimibe have been shown to impair HCV infection in vivo[50,52].
The importance of HCV entry owes to its critical role in the initiation of infection, the major target of immune responses and the determination of tissue and species tropism[24]. In recent years, substantial progress has been made to decipher the mystery of HCV entry. Current data support a complex interplay between the HCV-encoded glycoproteins E1 and E2 and four receptors - SR-BI, CD81, and tight junction proteins CLDN1 and OCLN in defining HCV entry. EGFR, NPC1L1 and TfR1 are important cofactors for HCV entry. HCV entry has been revealed as a highly complex process requiring orchestration of several cell surface molecules and regulation of multiple host kinases, which offer a multitude of promising targets for antivirals.
The ideal therapeutic regimen is thought to be an all-oral, IFN-free combination cocktail with pan-genotype coverage, minimal side effects and high virological cure rates in all patient groups[1]. Given the chronic nature and genetic diversity of HCV, targeting host entry factors with antibodies or small-molecule inhibitors could block the spread of DAA-resistant variants and disturb infection dynamics necessary to maintain chronic liver infection. The most advanced inhibitor of HCV entry is ITX 5061, a small molecule compound that was initially identified to promote HDL levels in animals and patients by targeting SRBI.
We are grateful to all of our colleagues for their help over the years and apologize if we have missed citing some articles due to space restrictions.
P- Reviewers: Dubuisson J, Song LT S- Editor: Cui XM L- Editor: Kerr C E- Editor: Zhang DN
| 1. | Scheel TK, Rice CM. Understanding the hepatitis C virus life cycle paves the way for highly effective therapies. Nat Med. 2013;19:837-849. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 438] [Cited by in F6Publishing: 408] [Article Influence: 37.1] [Reference Citation Analysis (0)] |
| 2. | Manns MP, Wedemeyer H, Cornberg M. Treating viral hepatitis C: efficacy, side effects, and complications. Gut. 2006;55:1350-1359. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 470] [Cited by in F6Publishing: 479] [Article Influence: 26.6] [Reference Citation Analysis (0)] |
| 3. | Jacobson IM, McHutchison JG, Dusheiko G, Di Bisceglie AM, Reddy KR, Bzowej NH, Marcellin P, Muir AJ, Ferenci P, Flisiak R. Telaprevir for previously untreated chronic hepatitis C virus infection. N Engl J Med. 2011;364:2405-2416. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1866] [Cited by in F6Publishing: 1835] [Article Influence: 141.2] [Reference Citation Analysis (0)] |
| 4. | Poordad F, McCone J, Bacon BR, Bruno S, Manns MP, Sulkowski MS, Jacobson IM, Reddy KR, Goodman ZD, Boparai N. Boceprevir for untreated chronic HCV genotype 1 infection. N Engl J Med. 2011;364:1195-1206. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1948] [Cited by in F6Publishing: 1951] [Article Influence: 150.1] [Reference Citation Analysis (0)] |
| 5. | Pawlotsky JM. Treatment failure and resistance with direct-acting antiviral drugs against hepatitis C virus. Hepatology. 2011;53:1742-1751. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 249] [Cited by in F6Publishing: 263] [Article Influence: 20.2] [Reference Citation Analysis (0)] |
| 6. | Bartosch B, Dubuisson J, Cosset FL. Infectious hepatitis C virus pseudo-particles containing functional E1-E2 envelope protein complexes. J Exp Med. 2003;197:633-642. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 885] [Cited by in F6Publishing: 868] [Article Influence: 41.3] [Reference Citation Analysis (0)] |
| 7. | Hsu M, Zhang J, Flint M, Logvinoff C, Cheng-Mayer C, Rice CM, McKeating JA. Hepatitis C virus glycoproteins mediate pH-dependent cell entry of pseudotyped retroviral particles. Proc Natl Acad Sci USA. 2003;100:7271-7276. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 655] [Cited by in F6Publishing: 640] [Article Influence: 30.5] [Reference Citation Analysis (0)] |
| 8. | Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, Murthy K, Habermann A, Kräusslich HG, Mizokami M. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med. 2005;11:791-796. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2309] [Cited by in F6Publishing: 2252] [Article Influence: 118.5] [Reference Citation Analysis (0)] |
| 9. | Lindenbach BD, Evans MJ, Syder AJ, Wölk B, Tellinghuisen TL, Liu CC, Maruyama T, Hynes RO, Burton DR, McKeating JA. Complete replication of hepatitis C virus in cell culture. Science. 2005;309:623-626. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1843] [Cited by in F6Publishing: 1819] [Article Influence: 95.7] [Reference Citation Analysis (0)] |
| 10. | Zhong J, Gastaminza P, Cheng G, Kapadia S, Kato T, Burton DR, Wieland SF, Uprichard SL, Wakita T, Chisari FV. Robust hepatitis C virus infection in vitro. Proc Natl Acad Sci USA. 2005;102:9294-9299. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1459] [Cited by in F6Publishing: 1449] [Article Influence: 76.3] [Reference Citation Analysis (0)] |
| 11. | Dorner M, Horwitz JA, Robbins JB, Barry WT, Feng Q, Mu K, Jones CT, Schoggins JW, Catanese MT, Burton DR. A genetically humanized mouse model for hepatitis C virus infection. Nature. 2011;474:208-211. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 293] [Cited by in F6Publishing: 285] [Article Influence: 21.9] [Reference Citation Analysis (0)] |
| 12. | Mercer DF, Schiller DE, Elliott JF, Douglas DN, Hao C, Rinfret A, Addison WR, Fischer KP, Churchill TA, Lakey JR. Hepatitis C virus replication in mice with chimeric human livers. Nat Med. 2001;7:927-933. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 693] [Cited by in F6Publishing: 672] [Article Influence: 29.2] [Reference Citation Analysis (0)] |
| 13. | Zhao X, Tang ZY, Klumpp B, Wolff-Vorbeck G, Barth H, Levy S, von Weizsäcker F, Blum HE, Baumert TF. Primary hepatocytes of Tupaia belangeri as a potential model for hepatitis C virus infection. J Clin Invest. 2002;109:221-232. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 39] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 14. | Tong Y, Zhu Y, Xia X, Liu Y, Feng Y, Hua X, Chen Z, Ding H, Gao L, Wang Y. Tupaia CD81, SR-BI, claudin-1, and occludin support hepatitis C virus infection. J Virol. 2011;85:2793-2802. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 62] [Cited by in F6Publishing: 69] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 15. | Meredith LW, Wilson GK, Fletcher NF, McKeating JA. Hepatitis C virus entry: beyond receptors. Rev Med Virol. 2012;22:182-193. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 72] [Cited by in F6Publishing: 76] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 16. | Zeisel MB, Felmlee DJ, Baumert TF. Hepatitis C virus entry. Curr Top Microbiol Immunol. 2013;369:87-112. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 39] [Cited by in F6Publishing: 78] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 17. | Gastaminza P, Dryden KA, Boyd B, Wood MR, Law M, Yeager M, Chisari FV. Ultrastructural and biophysical characterization of hepatitis C virus particles produced in cell culture. J Virol. 2010;84:10999-11009. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 166] [Cited by in F6Publishing: 160] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 18. | Vieyres G, Thomas X, Descamps V, Duverlie G, Patel AH, Dubuisson J. Characterization of the envelope glycoproteins associated with infectious hepatitis C virus. J Virol. 2010;84:10159-10168. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 168] [Cited by in F6Publishing: 169] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 19. | Wang W, Guan M, Liu Y, Xu Q, Peng H, Liu X, Tang Z, Zhu Y, Wu D, Ren H. Alanine scanning mutagenesis of hepatitis C virus E2 cysteine residues: Insights into E2 biogenesis and antigenicity. Virology. 2014;448:229-237. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 11] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 20. | Op De Beeck A, Voisset C, Bartosch B, Ciczora Y, Cocquerel L, Keck Z, Foung S, Cosset FL, Dubuisson J. Characterization of functional hepatitis C virus envelope glycoproteins. J Virol. 2004;78:2994-3002. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 171] [Cited by in F6Publishing: 181] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 21. | Tscherne DM, Jones CT, Evans MJ, Lindenbach BD, McKeating JA, Rice CM. Time- and temperature-dependent activation of hepatitis C virus for low-pH-triggered entry. J Virol. 2006;80:1734-1741. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 294] [Cited by in F6Publishing: 302] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 22. | Blanchard E, Belouzard S, Goueslain L, Wakita T, Dubuisson J, Wychowski C, Rouillé Y. Hepatitis C virus entry depends on clathrin-mediated endocytosis. J Virol. 2006;80:6964-6972. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 437] [Cited by in F6Publishing: 424] [Article Influence: 23.6] [Reference Citation Analysis (0)] |
| 23. | Ploss A, Evans MJ. Hepatitis C virus host cell entry. Curr Opin Virol. 2012;2:14-19. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 70] [Cited by in F6Publishing: 73] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 24. | Guan M, Wang W, Liu X, Tong Y, Liu Y, Ren H, Zhu S, Dubuisson J, Baumert TF, Zhu Y. Three different functional microdomains in the hepatitis C virus hypervariable region 1 (HVR1) mediate entry and immune evasion. J Biol Chem. 2012;287:35631-35645. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 44] [Cited by in F6Publishing: 41] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 25. | Kong L, Giang E, Nieusma T, Kadam RU, Cogburn KE, Hua Y, Dai X, Stanfield RL, Burton DR, Ward AB. Hepatitis C virus E2 envelope glycoprotein core structure. Science. 2013;342:1090-1094. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 314] [Cited by in F6Publishing: 311] [Article Influence: 28.3] [Reference Citation Analysis (0)] |
| 26. | Krey T, d’Alayer J, Kikuti CM, Saulnier A, Damier-Piolle L, Petitpas I, Johansson DX, Tawar RG, Baron B, Robert B. The disulfide bonds in glycoprotein E2 of hepatitis C virus reveal the tertiary organization of the molecule. PLoS Pathog. 2010;6:e1000762. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 185] [Cited by in F6Publishing: 189] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 27. | Lavillette D, Pécheur EI, Donot P, Fresquet J, Molle J, Corbau R, Dreux M, Penin F, Cosset FL. Characterization of fusion determinants points to the involvement of three discrete regions of both E1 and E2 glycoproteins in the membrane fusion process of hepatitis C virus. J Virol. 2007;81:8752-8765. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 133] [Cited by in F6Publishing: 139] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 28. | Li Y, Wang J, Kanai R, Modis Y. Crystal structure of glycoprotein E2 from bovine viral diarrhea virus. Proc Natl Acad Sci USA. 2013;110:6805-6810. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 90] [Cited by in F6Publishing: 99] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 29. | El Omari K, Iourin O, Harlos K, Grimes JM, Stuart DI. Structure of a pestivirus envelope glycoprotein E2 clarifies its role in cell entry. Cell Rep. 2013;3:30-35. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 114] [Cited by in F6Publishing: 114] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 30. | Wahid A, Helle F, Descamps V, Duverlie G, Penin F, Dubuisson J. Disulfide bonds in hepatitis C virus glycoprotein E1 control the assembly and entry functions of E2 glycoprotein. J Virol. 2013;87:1605-1617. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 49] [Cited by in F6Publishing: 45] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 31. | André P, Komurian-Pradel F, Deforges S, Perret M, Berland JL, Sodoyer M, Pol S, Bréchot C, Paranhos-Baccalà G, Lotteau V. Characterization of low- and very-low-density hepatitis C virus RNA-containing particles. J Virol. 2002;76:6919-6928. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 495] [Cited by in F6Publishing: 503] [Article Influence: 22.9] [Reference Citation Analysis (0)] |
| 32. | Dao Thi VL, Dreux M, Cosset FL. Scavenger receptor class B type I and the hypervariable region-1 of hepatitis C virus in cell entry and neutralisation. Expert Rev Mol Med. 2011;13:e13. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 34] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 33. | Gastaminza P, Cheng G, Wieland S, Zhong J, Liao W, Chisari FV. Cellular determinants of hepatitis C virus assembly, maturation, degradation, and secretion. J Virol. 2008;82:2120-2129. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 347] [Cited by in F6Publishing: 349] [Article Influence: 20.5] [Reference Citation Analysis (0)] |
| 34. | Nielsen SU, Bassendine MF, Burt AD, Martin C, Pumeechockchai W, Toms GL. Association between hepatitis C virus and very-low-density lipoprotein (VLDL)/LDL analyzed in iodixanol density gradients. J Virol. 2006;80:2418-2428. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 262] [Cited by in F6Publishing: 256] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 35. | André P, Perlemuter G, Budkowska A, Bréchot C, Lotteau V. Hepatitis C virus particles and lipoprotein metabolism. Semin Liver Dis. 2005;25:93-104. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 129] [Cited by in F6Publishing: 129] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 36. | Icard V, Diaz O, Scholtes C, Perrin-Cocon L, Ramière C, Bartenschlager R, Penin F, Lotteau V, André P. Secretion of hepatitis C virus envelope glycoproteins depends on assembly of apolipoprotein B positive lipoproteins. PLoS One. 2009;4:e4233. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 104] [Cited by in F6Publishing: 111] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 37. | Bartenschlager R, Penin F, Lohmann V, André P. Assembly of infectious hepatitis C virus particles. Trends Microbiol. 2011;19:95-103. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 320] [Cited by in F6Publishing: 329] [Article Influence: 23.5] [Reference Citation Analysis (0)] |
| 38. | Meunier JC, Russell RS, Engle RE, Faulk KN, Purcell RH, Emerson SU. Apolipoprotein c1 association with hepatitis C virus. J Virol. 2008;82:9647-9656. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 108] [Cited by in F6Publishing: 110] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 39. | Felmlee DJ, Hafirassou ML, Lefevre M, Baumert TF, Schuster C. Hepatitis C virus, cholesterol and lipoproteins--impact for the viral life cycle and pathogenesis of liver disease. Viruses. 2013;5:1292-1324. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 102] [Cited by in F6Publishing: 106] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 40. | Koutsoudakis G, Kaul A, Steinmann E, Kallis S, Lohmann V, Pietschmann T, Bartenschlager R. Characterization of the early steps of hepatitis C virus infection by using luciferase reporter viruses. J Virol. 2006;80:5308-5320. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 321] [Cited by in F6Publishing: 332] [Article Influence: 19.5] [Reference Citation Analysis (0)] |
| 41. | Barth H, Schafer C, Adah MI, Zhang F, Linhardt RJ, Toyoda H, Kinoshita-Toyoda A, Toida T, Van Kuppevelt TH, Depla E. Cellular binding of hepatitis C virus envelope glycoprotein E2 requires cell surface heparan sulfate. J Biol Chem. 2003;278:41003-41012. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 356] [Cited by in F6Publishing: 368] [Article Influence: 17.5] [Reference Citation Analysis (0)] |
| 42. | Molina S, Castet V, Fournier-Wirth C, Pichard-Garcia L, Avner R, Harats D, Roitelman J, Barbaras R, Graber P, Ghersa P. The low-density lipoprotein receptor plays a role in the infection of primary human hepatocytes by hepatitis C virus. J Hepatol. 2007;46:411-419. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 214] [Cited by in F6Publishing: 214] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 43. | Agnello V, Abel G, Elfahal M, Knight GB, Zhang QX. Hepatitis C virus and other flaviviridae viruses enter cells via low density lipoprotein receptor. Proc Natl Acad Sci USA. 1999;96:12766-12771. [PubMed] [Cited in This Article: ] |
| 44. | Monazahian M, Böhme I, Bonk S, Koch A, Scholz C, Grethe S, Thomssen R. Low density lipoprotein receptor as a candidate receptor for hepatitis C virus. J Med Virol. 1999;57:223-229. [PubMed] [Cited in This Article: ] |
| 45. | Owen DM, Huang H, Ye J, Gale M. Apolipoprotein E on hepatitis C virion facilitates infection through interaction with low-density lipoprotein receptor. Virology. 2009;394:99-108. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 166] [Cited by in F6Publishing: 170] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 46. | Scarselli E, Ansuini H, Cerino R, Roccasecca RM, Acali S, Filocamo G, Traboni C, Nicosia A, Cortese R, Vitelli A. The human scavenger receptor class B type I is a novel candidate receptor for the hepatitis C virus. EMBO J. 2002;21:5017-5025. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 888] [Cited by in F6Publishing: 868] [Article Influence: 39.5] [Reference Citation Analysis (0)] |
| 47. | Pileri P, Uematsu Y, Campagnoli S, Galli G, Falugi F, Petracca R, Weiner AJ, Houghton M, Rosa D, Grandi G. Binding of hepatitis C virus to CD81. Science. 1998;282:938-941. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1572] [Cited by in F6Publishing: 1532] [Article Influence: 58.9] [Reference Citation Analysis (0)] |
| 48. | Evans MJ, von Hahn T, Tscherne DM, Syder AJ, Panis M, Wölk B, Hatziioannou T, McKeating JA, Bieniasz PD, Rice CM. Claudin-1 is a hepatitis C virus co-receptor required for a late step in entry. Nature. 2007;446:801-805. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 930] [Cited by in F6Publishing: 923] [Article Influence: 54.3] [Reference Citation Analysis (0)] |
| 49. | Ploss A, Evans MJ, Gaysinskaya VA, Panis M, You H, de Jong YP, Rice CM. Human occludin is a hepatitis C virus entry factor required for infection of mouse cells. Nature. 2009;457:882-886. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 715] [Cited by in F6Publishing: 705] [Article Influence: 47.0] [Reference Citation Analysis (0)] |
| 50. | Lupberger J, Zeisel MB, Xiao F, Thumann C, Fofana I, Zona L, Davis C, Mee CJ, Turek M, Gorke S. EGFR and EphA2 are host factors for hepatitis C virus entry and possible targets for antiviral therapy. Nat Med. 2011;17:589-595. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 574] [Cited by in F6Publishing: 555] [Article Influence: 42.7] [Reference Citation Analysis (0)] |
| 51. | Martin DN, Uprichard SL. Identification of transferrin receptor 1 as a hepatitis C virus entry factor. Proc Natl Acad Sci USA. 2013;110:10777-10782. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 157] [Cited by in F6Publishing: 163] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 52. | Sainz B, Barretto N, Martin DN, Hiraga N, Imamura M, Hussain S, Marsh KA, Yu X, Chayama K, Alrefai WA. Identification of the Niemann-Pick C1-like 1 cholesterol absorption receptor as a new hepatitis C virus entry factor. Nat Med. 2012;18:281-285. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 349] [Cited by in F6Publishing: 348] [Article Influence: 29.0] [Reference Citation Analysis (0)] |
| 53. | Lozach PY, Lortat-Jacob H, de Lacroix de Lavalette A, Staropoli I, Foung S, Amara A, Houles C, Fieschi F, Schwartz O, Virelizier JL. DC-SIGN and L-SIGN are high affinity binding receptors for hepatitis C virus glycoprotein E2. J Biol Chem. 2003;278:20358-20366. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 281] [Cited by in F6Publishing: 285] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 54. | Pöhlmann S, Zhang J, Baribaud F, Chen Z, Leslie GJ, Lin G, Granelli-Piperno A, Doms RW, Rice CM, McKeating JA. Hepatitis C virus glycoproteins interact with DC-SIGN and DC-SIGNR. J Virol. 2003;77:4070-4080. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 296] [Cited by in F6Publishing: 310] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 55. | Barth H, Schnober EK, Zhang F, Linhardt RJ, Depla E, Boson B, Cosset FL, Patel AH, Blum HE, Baumert TF. Viral and cellular determinants of the hepatitis C virus envelope-heparan sulfate interaction. J Virol. 2006;80:10579-10590. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 136] [Cited by in F6Publishing: 146] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 56. | Jiang J, Cun W, Wu X, Shi Q, Tang H, Luo G. Hepatitis C virus attachment mediated by apolipoprotein E binding to cell surface heparan sulfate. J Virol. 2012;86:7256-7267. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 126] [Cited by in F6Publishing: 126] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 57. | Albecka A, Belouzard S, Op de Beeck A, Descamps V, Goueslain L, Bertrand-Michel J, Tercé F, Duverlie G, Rouillé Y, Dubuisson J. Role of low-density lipoprotein receptor in the hepatitis C virus life cycle. Hepatology. 2012;55:998-1007. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 123] [Cited by in F6Publishing: 124] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 58. | Rhainds D, Brodeur M, Lapointe J, Charpentier D, Falstrault L, Brissette L. The role of human and mouse hepatic scavenger receptor class B type I (SR-BI) in the selective uptake of low-density lipoprotein-cholesteryl esters. Biochemistry. 2003;42:7527-7538. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 47] [Cited by in F6Publishing: 49] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 59. | Van Eck M, Hoekstra M, Out R, Bos IS, Kruijt JK, Hildebrand RB, Van Berkel TJ. Scavenger receptor BI facilitates the metabolism of VLDL lipoproteins in vivo. J Lipid Res. 2008;49:136-146. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 74] [Cited by in F6Publishing: 79] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 60. | Rhainds D, Brissette L. The role of scavenger receptor class B type I (SR-BI) in lipid trafficking. defining the rules for lipid traders. Int J Biochem Cell Biol. 2004;36:39-77. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 120] [Cited by in F6Publishing: 158] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 61. | Bartosch B, Verney G, Dreux M, Donot P, Morice Y, Penin F, Pawlotsky JM, Lavillette D, Cosset FL. An interplay between hypervariable region 1 of the hepatitis C virus E2 glycoprotein, the scavenger receptor BI, and high-density lipoprotein promotes both enhancement of infection and protection against neutralizing antibodies. J Virol. 2005;79:8217-8229. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 227] [Cited by in F6Publishing: 227] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 62. | Dreux M, Dao Thi VL, Fresquet J, Guérin M, Julia Z, Verney G, Durantel D, Zoulim F, Lavillette D, Cosset FL. Receptor complementation and mutagenesis reveal SR-BI as an essential HCV entry factor and functionally imply its intra- and extra-cellular domains. PLoS Pathog. 2009;5:e1000310. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 98] [Cited by in F6Publishing: 102] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 63. | Dao Thi VL, Granier C, Zeisel MB, Guérin M, Mancip J, Granio O, Penin F, Lavillette D, Bartenschlager R, Baumert TF. Characterization of hepatitis C virus particle subpopulations reveals multiple usage of the scavenger receptor BI for entry steps. J Biol Chem. 2012;287:31242-31257. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 100] [Cited by in F6Publishing: 97] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 64. | Maillard P, Huby T, Andréo U, Moreau M, Chapman J, Budkowska A. The interaction of natural hepatitis C virus with human scavenger receptor SR-BI/Cla1 is mediated by ApoB-containing lipoproteins. FASEB J. 2006;20:735-737. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 109] [Cited by in F6Publishing: 118] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 65. | Zahid MN, Turek M, Xiao F, Thi VL, Guérin M, Fofana I, Bachellier P, Thompson J, Delang L, Neyts J. The postbinding activity of scavenger receptor class B type I mediates initiation of hepatitis C virus infection and viral dissemination. Hepatology. 2013;57:492-504. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 61] [Cited by in F6Publishing: 64] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 66. | Flint M, von Hahn T, Zhang J, Farquhar M, Jones CT, Balfe P, Rice CM, McKeating JA. Diverse CD81 proteins support hepatitis C virus infection. J Virol. 2006;80:11331-11342. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 124] [Cited by in F6Publishing: 133] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 67. | Sharma NR, Mateu G, Dreux M, Grakoui A, Cosset FL, Melikyan GB. Hepatitis C virus is primed by CD81 protein for low pH-dependent fusion. J Biol Chem. 2011;286:30361-30376. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 75] [Cited by in F6Publishing: 79] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 68. | Brazzoli M, Bianchi A, Filippini S, Weiner A, Zhu Q, Pizza M, Crotta S. CD81 is a central regulator of cellular events required for hepatitis C virus infection of human hepatocytes. J Virol. 2008;82:8316-8329. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 142] [Cited by in F6Publishing: 142] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 69. | Rocha-Perugini V, Montpellier C, Delgrange D, Wychowski C, Helle F, Pillez A, Drobecq H, Le Naour F, Charrin S, Levy S. The CD81 partner EWI-2wint inhibits hepatitis C virus entry. PLoS One. 2008;3:e1866. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 88] [Cited by in F6Publishing: 91] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 70. | Montpellier C, Tews BA, Poitrimole J, Rocha-Perugini V, D’Arienzo V, Potel J, Zhang XA, Rubinstein E, Dubuisson J, Cocquerel L. Interacting regions of CD81 and two of its partners, EWI-2 and EWI-2wint, and their effect on hepatitis C virus infection. J Biol Chem. 2011;286:13954-13965. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 45] [Cited by in F6Publishing: 46] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 71. | Zhu YZ, Luo Y, Cao MM, Liu Y, Liu XQ, Wang W, Wu DG, Guan M, Xu QQ, Ren H. Significance of palmitoylation of CD81 on its association with tetraspanin-enriched microdomains and mediating hepatitis C virus cell entry. Virology. 2012;429:112-123. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 23] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 72. | Zheng A, Yuan F, Li Y, Zhu F, Hou P, Li J, Song X, Ding M, Deng H. Claudin-6 and claudin-9 function as additional coreceptors for hepatitis C virus. J Virol. 2007;81:12465-12471. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 156] [Cited by in F6Publishing: 166] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 73. | Krieger SE, Zeisel MB, Davis C, Thumann C, Harris HJ, Schnober EK, Mee C, Soulier E, Royer C, Lambotin M. Inhibition of hepatitis C virus infection by anti-claudin-1 antibodies is mediated by neutralization of E2-CD81-claudin-1 associations. Hepatology. 2010;51:1144-1157. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 137] [Cited by in F6Publishing: 135] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 74. | Harris HJ, Farquhar MJ, Mee CJ, Davis C, Reynolds GM, Jennings A, Hu K, Yuan F, Deng H, Hubscher SG. CD81 and claudin 1 coreceptor association: role in hepatitis C virus entry. J Virol. 2008;82:5007-5020. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 152] [Cited by in F6Publishing: 154] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 75. | Mee CJ, Harris HJ, Farquhar MJ, Wilson G, Reynolds G, Davis C, van IJzendoorn SC, Balfe P, McKeating JA. Polarization restricts hepatitis C virus entry into HepG2 hepatoma cells. J Virol. 2009;83:6211-6221. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 104] [Cited by in F6Publishing: 106] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 76. | Schneider MR, Wolf E. The epidermal growth factor receptor ligands at a glance. J Cell Physiol. 2009;218:460-466. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 291] [Cited by in F6Publishing: 301] [Article Influence: 20.1] [Reference Citation Analysis (0)] |
| 77. | Lackmann M, Boyd AW. Eph, a protein family coming of age: more confusion, insight, or complexity? Sci Signal. 2008;1:re2. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 118] [Cited by in F6Publishing: 125] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 78. | Zona L, Lupberger J, Sidahmed-Adrar N, Thumann C, Harris HJ, Barnes A, Florentin J, Tawar RG, Xiao F, Turek M. HRas signal transduction promotes hepatitis C virus cell entry by triggering assembly of the host tetraspanin receptor complex. Cell Host Microbe. 2013;13:302-313. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 126] [Cited by in F6Publishing: 129] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 79. | Furuse M, Hirase T, Itoh M, Nagafuchi A, Yonemura S, Tsukita S, Tsukita S. Occludin: a novel integral membrane protein localizing at tight junctions. J Cell Biol. 1993;123:1777-1788. [PubMed] [Cited in This Article: ] |
| 80. | Liu S, Yang W, Shen L, Turner JR, Coyne CB, Wang T. Tight junction proteins claudin-1 and occludin control hepatitis C virus entry and are downregulated during infection to prevent superinfection. J Virol. 2009;83:2011-2014. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 264] [Cited by in F6Publishing: 262] [Article Influence: 16.4] [Reference Citation Analysis (0)] |
| 81. | Zeisel MB, Fofana I, Fafi-Kremer S, Baumert TF. Hepatitis C virus entry into hepatocytes: molecular mechanisms and targets for antiviral therapies. J Hepatol. 2011;54:566-576. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 136] [Cited by in F6Publishing: 138] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 82. | Farquhar MJ, Hu K, Harris HJ, Davis C, Brimacombe CL, Fletcher SJ, Baumert TF, Rappoport JZ, Balfe P, McKeating JA. Hepatitis C virus induces CD81 and claudin-1 endocytosis. J Virol. 2012;86:4305-4316. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 99] [Cited by in F6Publishing: 96] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 83. | Haid S, Pietschmann T, Pécheur EI. Low pH-dependent hepatitis C virus membrane fusion depends on E2 integrity, target lipid composition, and density of virus particles. J Biol Chem. 2009;284:17657-17667. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 71] [Cited by in F6Publishing: 75] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 84. | Kielian M, Rey FA. Virus membrane-fusion proteins: more than one way to make a hairpin. Nat Rev Microbiol. 2006;4:67-76. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 421] [Cited by in F6Publishing: 427] [Article Influence: 23.7] [Reference Citation Analysis (0)] |
| 85. | Li HF, Huang CH, Ai LS, Chuang CK, Chen SS. Mutagenesis of the fusion peptide-like domain of hepatitis C virus E1 glycoprotein: involvement in cell fusion and virus entry. J Biomed Sci. 2009;16:89. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 39] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 86. | Drummer HE, Boo I, Poumbourios P. Mutagenesis of a conserved fusion peptide-like motif and membrane-proximal heptad-repeat region of hepatitis C virus glycoprotein E1. J Gen Virol. 2007;88:1144-1148. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 71] [Cited by in F6Publishing: 78] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 87. | Stiasny K, Fritz R, Pangerl K, Heinz FX. Molecular mechanisms of flavivirus membrane fusion. Amino Acids. 2011;41:1159-1163. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 77] [Cited by in F6Publishing: 81] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 88. | Mukhopadhyay S, Kuhn RJ, Rossmann MG. A structural perspective of the flavivirus life cycle. Nat Rev Microbiol. 2005;3:13-22. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 828] [Cited by in F6Publishing: 864] [Article Influence: 45.5] [Reference Citation Analysis (0)] |
| 89. | Corver J, Ortiz A, Allison SL, Schalich J, Heinz FX, Wilschut J. Membrane fusion activity of tick-borne encephalitis virus and recombinant subviral particles in a liposomal model system. Virology. 2000;269:37-46. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 107] [Cited by in F6Publishing: 105] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 90. | Moesker B, Rodenhuis-Zybert IA, Meijerhof T, Wilschut J, Smit JM. Characterization of the functional requirements of West Nile virus membrane fusion. J Gen Virol. 2010;91:389-393. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 39] [Cited by in F6Publishing: 45] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 91. | Stiasny K, Koessl C, Heinz FX. Involvement of lipids in different steps of the flavivirus fusion mechanism. J Virol. 2003;77:7856-7862. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 75] [Cited by in F6Publishing: 74] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 92. | Lupberger J, Felmlee DJ, Baumert TF. Cholesterol uptake and hepatitis C virus entry. J Hepatol. 2012;57:215-217. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 7] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 93. | Kim S, Ishida H, Yamane D, Yi M, Swinney DC, Foung S, Lemon SM. Contrasting roles of mitogen-activated protein kinases in cellular entry and replication of hepatitis C virus: MKNK1 facilitates cell entry. J Virol. 2013;87:4214-4224. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 27] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 94. | Marsh M, Helenius A. Virus entry: open sesame. Cell. 2006;124:729-740. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 863] [Cited by in F6Publishing: 916] [Article Influence: 50.9] [Reference Citation Analysis (0)] |
| 95. | Harris HJ, Davis C, Mullins JG, Hu K, Goodall M, Farquhar MJ, Mee CJ, McCaffrey K, Young S, Drummer H. Claudin association with CD81 defines hepatitis C virus entry. J Biol Chem. 2010;285:21092-21102. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 165] [Cited by in F6Publishing: 160] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 96. | Farquhar MJ, Harris HJ, Diskar M, Jones S, Mee CJ, Nielsen SU, Brimacombe CL, Molina S, Toms GL, Maurel P. Protein kinase A-dependent step(s) in hepatitis C virus entry and infectivity. J Virol. 2008;82:8797-8811. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 75] [Cited by in F6Publishing: 77] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 97. | Lee CJ, Liao CL, Lin YL. Flavivirus activates phosphatidylinositol 3-kinase signaling to block caspase-dependent apoptotic cell death at the early stage of virus infection. J Virol. 2005;79:8388-8399. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 160] [Cited by in F6Publishing: 155] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 98. | Zhu YZ, Cao MM, Wang WB, Wang W, Ren H, Zhao P, Qi ZT. Association of heat-shock protein 70 with lipid rafts is required for Japanese encephalitis virus infection in Huh7 cells. J Gen Virol. 2012;93:61-71. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 54] [Cited by in F6Publishing: 58] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 99. | Liu Z, Tian Y, Machida K, Lai MM, Luo G, Foung SK, Ou JH. Transient activation of the PI3K-AKT pathway by hepatitis C virus to enhance viral entry. J Biol Chem. 2012;287:41922-41930. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 67] [Cited by in F6Publishing: 68] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 100. | Trotard M, Lepère-Douard C, Régeard M, Piquet-Pellorce C, Lavillette D, Cosset FL, Gripon P, Le Seyec J. Kinases required in hepatitis C virus entry and replication highlighted by small interference RNA screening. FASEB J. 2009;23:3780-3789. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 120] [Cited by in F6Publishing: 127] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 101. | Berger KL, Cooper JD, Heaton NS, Yoon R, Oakland TE, Jordan TX, Mateu G, Grakoui A, Randall G. Roles for endocytic trafficking and phosphatidylinositol 4-kinase III alpha in hepatitis C virus replication. Proc Natl Acad Sci USA. 2009;106:7577-7582. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 275] [Cited by in F6Publishing: 278] [Article Influence: 18.5] [Reference Citation Analysis (0)] |
| 102. | Owsianka A, Tarr AW, Juttla VS, Lavillette D, Bartosch B, Cosset FL, Ball JK, Patel AH. Monoclonal antibody AP33 defines a broadly neutralizing epitope on the hepatitis C virus E2 envelope glycoprotein. J Virol. 2005;79:11095-11104. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 232] [Cited by in F6Publishing: 234] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 103. | Davis GL, Nelson DR, Terrault N, Pruett TL, Schiano TD, Fletcher CV, Sapan CV, Riser LN, Li Y, Whitley RJ. A randomized, open-label study to evaluate the safety and pharmacokinetics of human hepatitis C immune globulin (Civacir) in liver transplant recipients. Liver Transpl. 2005;11:941-949. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 136] [Cited by in F6Publishing: 138] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 104. | Edwards VC, Tarr AW, Urbanowicz RA, Ball JK. The role of neutralizing antibodies in hepatitis C virus infection. J Gen Virol. 2012;93:1-19. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 45] [Cited by in F6Publishing: 47] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 105. | Wahid A, Dubuisson J. Virus-neutralizing antibodies to hepatitis C virus. J Viral Hepat. 2013;20:369-376. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 45] [Cited by in F6Publishing: 48] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 106. | Helle F, Wychowski C, Vu-Dac N, Gustafson KR, Voisset C, Dubuisson J. Cyanovirin-N inhibits hepatitis C virus entry by binding to envelope protein glycans. J Biol Chem. 2006;281:25177-25183. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 129] [Cited by in F6Publishing: 129] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 107. | Meuleman P, Albecka A, Belouzard S, Vercauteren K, Verhoye L, Wychowski C, Leroux-Roels G, Palmer KE, Dubuisson J. Griffithsin has antiviral activity against hepatitis C virus. Antimicrob Agents Chemother. 2011;55:5159-5167. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 109] [Cited by in F6Publishing: 118] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 108. | Chang KS, Jiang J, Cai Z, Luo G. Human apolipoprotein e is required for infectivity and production of hepatitis C virus in cell culture. J Virol. 2007;81:13783-13793. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 328] [Cited by in F6Publishing: 333] [Article Influence: 19.6] [Reference Citation Analysis (0)] |
| 109. | Ciesek S, von Hahn T, Colpitts CC, Schang LM, Friesland M, Steinmann J, Manns MP, Ott M, Wedemeyer H, Meuleman P. The green tea polyphenol, epigallocatechin-3-gallate, inhibits hepatitis C virus entry. Hepatology. 2011;54:1947-1955. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 214] [Cited by in F6Publishing: 207] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 110. | Calland N, Albecka A, Belouzard S, Wychowski C, Duverlie G, Descamps V, Hober D, Dubuisson J, Rouillé Y, Séron K. (-)-Epigallocatechin-3-gallate is a new inhibitor of hepatitis C virus entry. Hepatology. 2012;55:720-729. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 190] [Cited by in F6Publishing: 184] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 111. | Yu F, Wang Q, Zhang Z, Peng Y, Qiu Y, Shi Y, Zheng Y, Xiao S, Wang H, Huang X. Development of oleanane-type triterpenes as a new class of HCV entry inhibitors. J Med Chem. 2013;56:4300-4319. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 71] [Cited by in F6Publishing: 78] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 112. | Lemon SM, McKeating JA, Pietschmann T, Frick DN, Glenn JS, Tellinghuisen TL, Symons J, Furman PA. Development of novel therapies for hepatitis C. Antiviral Res. 2010;86:79-92. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 60] [Cited by in F6Publishing: 53] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 113. | Meuleman P, Catanese MT, Verhoye L, Desombere I, Farhoudi A, Jones CT, Sheahan T, Grzyb K, Cortese R, Rice CM. A human monoclonal antibody targeting scavenger receptor class B type I precludes hepatitis C virus infection and viral spread in vitro and in vivo. Hepatology. 2012;55:364-372. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 106] [Cited by in F6Publishing: 105] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 114. | Meuleman P, Hesselgesser J, Paulson M, Vanwolleghem T, Desombere I, Reiser H, Leroux-Roels G. Anti-CD81 antibodies can prevent a hepatitis C virus infection in vivo. Hepatology. 2008;48:1761-1768. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 179] [Cited by in F6Publishing: 176] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 115. | Fofana I, Krieger SE, Grunert F, Glauben S, Xiao F, Fafi-Kremer S, Soulier E, Royer C, Thumann C, Mee CJ. Monoclonal anti-claudin 1 antibodies prevent hepatitis C virus infection of primary human hepatocytes. Gastroenterology. 2010;139:953-964, 964.e1-4. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 137] [Cited by in F6Publishing: 139] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 116. | Si Y, Liu S, Liu X, Jacobs JL, Cheng M, Niu Y, Jin Q, Wang T, Yang W. A human claudin-1-derived peptide inhibits hepatitis C virus entry. Hepatology. 2012;56:507-515. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 41] [Cited by in F6Publishing: 43] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 117. | Syder AJ, Lee H, Zeisel MB, Grove J, Soulier E, Macdonald J, Chow S, Chang J, Baumert TF, McKeating JA. Small molecule scavenger receptor BI antagonists are potent HCV entry inhibitors. J Hepatol. 2011;54:48-55. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 107] [Cited by in F6Publishing: 108] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 118. | Matsumura T, Hu Z, Kato T, Dreux M, Zhang YY, Imamura M, Hiraga N, Juteau JM, Cosset FL, Chayama K. Amphipathic DNA polymers inhibit hepatitis C virus infection by blocking viral entry. Gastroenterology. 2009;137:673-681. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 64] [Cited by in F6Publishing: 70] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 119. | Gastaminza P, Whitten-Bauer C, Chisari FV. Unbiased probing of the entire hepatitis C virus life cycle identifies clinical compounds that target multiple aspects of the infection. Proc Natl Acad Sci USA. 2010;107:291-296. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 92] [Cited by in F6Publishing: 97] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 120. | Chockalingam K, Simeon RL, Rice CM, Chen Z. A cell protection screen reveals potent inhibitors of multiple stages of the hepatitis C virus life cycle. Proc Natl Acad Sci USA. 2010;107:3764-3769. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 59] [Cited by in F6Publishing: 65] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 121. | Liu R, Tewari M, Kong R, Zhang R, Ingravallo P, Ralston R. A peptide derived from hepatitis C virus E2 envelope protein inhibits a post-binding step in HCV entry. Antiviral Res. 2010;86:172-179. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 18] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 122. | Blaising J, Lévy PL, Polyak SJ, Stanifer M, Boulant S, Pécheur EI. Arbidol inhibits viral entry by interfering with clathrin-dependent trafficking. Antiviral Res. 2013;100:215-219. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 57] [Cited by in F6Publishing: 57] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 123. | Murakami Y, Fukasawa M, Kaneko Y, Suzuki T, Wakita T, Fukazawa H. Selective estrogen receptor modulators inhibit hepatitis C virus infection at multiple steps of the virus life cycle. Microbes Infect. 2013;15:45-55. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 40] [Cited by in F6Publishing: 34] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 124. | Cui HK, Qing J, Guo Y, Wang YJ, Cui LJ, He TH, Zhang L, Liu L. Stapled peptide-based membrane fusion inhibitors of hepatitis C virus. Bioorg Med Chem. 2013;21:3547-3554. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 41] [Cited by in F6Publishing: 48] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 125. | Anggakusuma , Colpitts CC, Schang LM, Rachmawati H, Frentzen A, Pfaender S, Behrendt P, Brown RJ, Bankwitz D, Steinmann J. Turmeric curcumin inhibits entry of all hepatitis C virus genotypes into human liver cells. Gut. 2013;Epub ahead of print. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 129] [Cited by in F6Publishing: 119] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 126. | Chamoun-Emanuelli AM, Pecheur EI, Simeon RL, Huang D, Cremer PS, Chen Z. Phenothiazines inhibit hepatitis C virus entry, likely by increasing the fluidity of cholesterol-rich membranes. Antimicrob Agents Chemother. 2013;57:2571-2581. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 38] [Cited by in F6Publishing: 43] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 127. | Vausselin T, Calland N, Belouzard S, Descamps V, Douam F, Helle F, François C, Lavillette D, Duverlie G, Wahid A. The antimalarial ferroquine is an inhibitor of hepatitis C virus. Hepatology. 2013;58:86-97. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 40] [Cited by in F6Publishing: 42] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 128. | Colpitts CC, Ustinov AV, Epand RF, Epand RM, Korshun VA, Schang LM. 5-(Perylen-3-yl)ethynyl-arabino-uridine (aUY11), an arabino-based rigid amphipathic fusion inhibitor, targets virion envelope lipids to inhibit fusion of influenza virus, hepatitis C virus, and other enveloped viruses. J Virol. 2013;87:3640-3654. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 49] [Cited by in F6Publishing: 60] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 129. | Bush CO, Pokrovskii MV, Saito R, Morganelli P, Canales E, Clarke MO, Lazerwith SE, Golde J, Reid BG, Babaoglu K. A small-molecule inhibitor of hepatitis C virus infectivity. Antimicrob Agents Chemother. 2014;58:386-396. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 19] [Cited by in F6Publishing: 17] [Article Influence: 1.5] [Reference Citation Analysis (0)] |