Copyright
©2013 Baishideng Publishing Group Co.
World J Gastroenterol. Feb 21, 2013; 19(7): 1056-1067
Published online Feb 21, 2013. doi: 10.3748/wjg.v19.i7.1056
Published online Feb 21, 2013. doi: 10.3748/wjg.v19.i7.1056
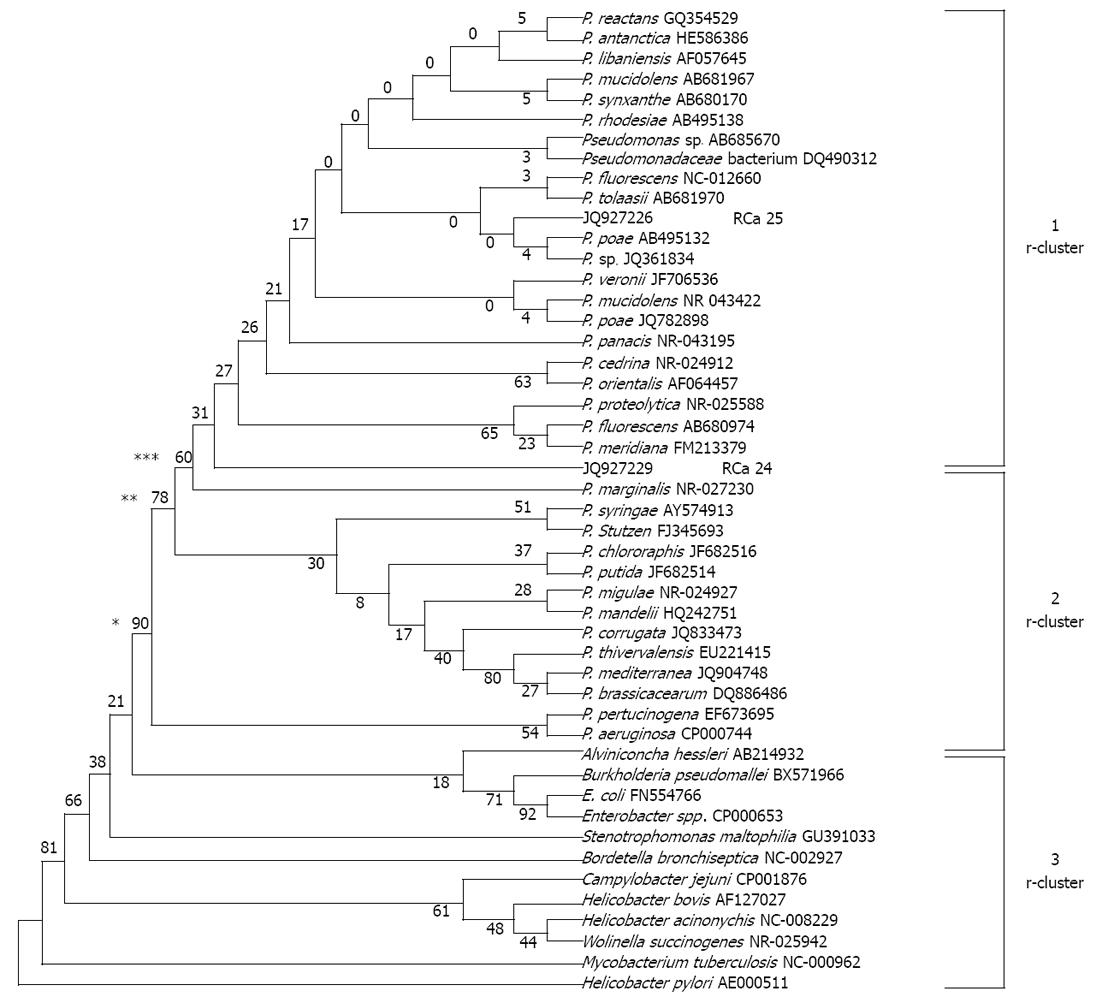
Figure 4 Phylogenetic affiliation of the Pseudomonas fluorescens-like isolates (n = 2; JQ927226 and JQ927227).
The unrooted tree was generated using unweighted pair group method with arithmetic mean from evolutionary distance computed with bootstrap test of phylogeny using MEGA version 4 by aligning published sequences from Genbank of 16S rRNA genes from 34 reference strains representative of the principal Pseudomonas (P.) phyla (accession number followed by species name in parentheses). For checking relatedness with other genera, we included 16S rRNA gene sequences from GenBank of 12 bacterial pathogens namely Stenotrophomonas maltophilia, Bordetella bronchiseptica, Wolinella succinogenes, Helicobacter pylori, Helicobacter acinonychis, Escherichia coli (E. coli), Enterobacter spp., Campylobacter jejuni, Burkholderia pseudomallei, Alviniconcha hessleri, Helicobacter bovis and Mycobacterium tuberculosis. Branches found by maximum likelihood are labeled with asterisks: one asterisk if bootstrap values = 90%, two asterisks if = 78% and three asterisks if = 60%.
-
Citation: Patel SK, Pratap CB, Verma AK, Jain AK, Dixit VK, Nath G.
Pseudomonas fluorescens -like bacteria from the stomach: A microbiological and molecular study. World J Gastroenterol 2013; 19(7): 1056-1067 - URL: https://www.wjgnet.com/1007-9327/full/v19/i7/1056.htm
- DOI: https://dx.doi.org/10.3748/wjg.v19.i7.1056