Copyright
©2013 Baishideng Publishing Group Co.
World J Gastroenterol. Sep 21, 2013; 19(35): 5813-5827
Published online Sep 21, 2013. doi: 10.3748/wjg.v19.i35.5813
Published online Sep 21, 2013. doi: 10.3748/wjg.v19.i35.5813
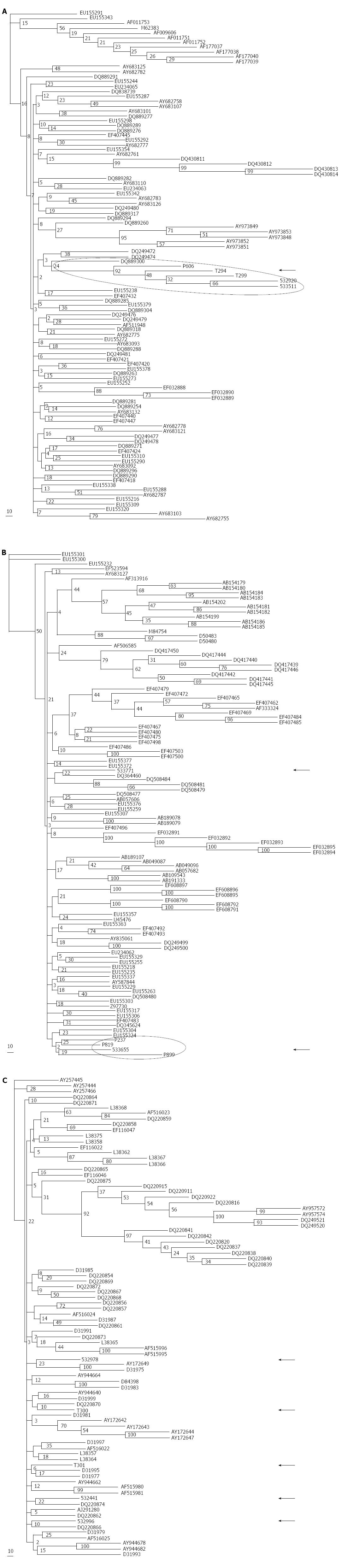
Figure 2 Phylogenetic trees performed by the Neighbor-Joining method.
NS5B sequences obtained from the volunteers whose respective hepatitis C virus (HCV) isolates had been classified as HCV-1a (n = 5), HCV-1b (n = 5) or HCV-2c (n = 5) were respectively analyzed in panels A, B and C together with 100 HCV-1a, 100 HCV-1b or 81 HCV-2c sequences downloaded from the GenBank. The respective consensus trees are shown after analyzing 100 replicate trees for each HCV subtype. A distance scale (in nucleotide substitutions per position) is shown in each panel. Arrows indicate sequences obtained from Argentinian volunteers.
- Citation: del Pino N, Oubiña JR, Rodríguez-Frías F, Esteban JI, Buti M, Otero T, Gregori J, García-Cehic D, Camos S, Cubero M, Casillas R, Guàrdia J, Esteban R, Quer J. Molecular epidemiology and putative origin of hepatitis C virus in random volunteers from Argentina. World J Gastroenterol 2013; 19(35): 5813-5827
- URL: https://www.wjgnet.com/1007-9327/full/v19/i35/5813.htm
- DOI: https://dx.doi.org/10.3748/wjg.v19.i35.5813