Copyright
©2013 Baishideng Publishing Group Co.
World J Gastroenterol. Sep 21, 2013; 19(35): 5813-5827
Published online Sep 21, 2013. doi: 10.3748/wjg.v19.i35.5813
Published online Sep 21, 2013. doi: 10.3748/wjg.v19.i35.5813
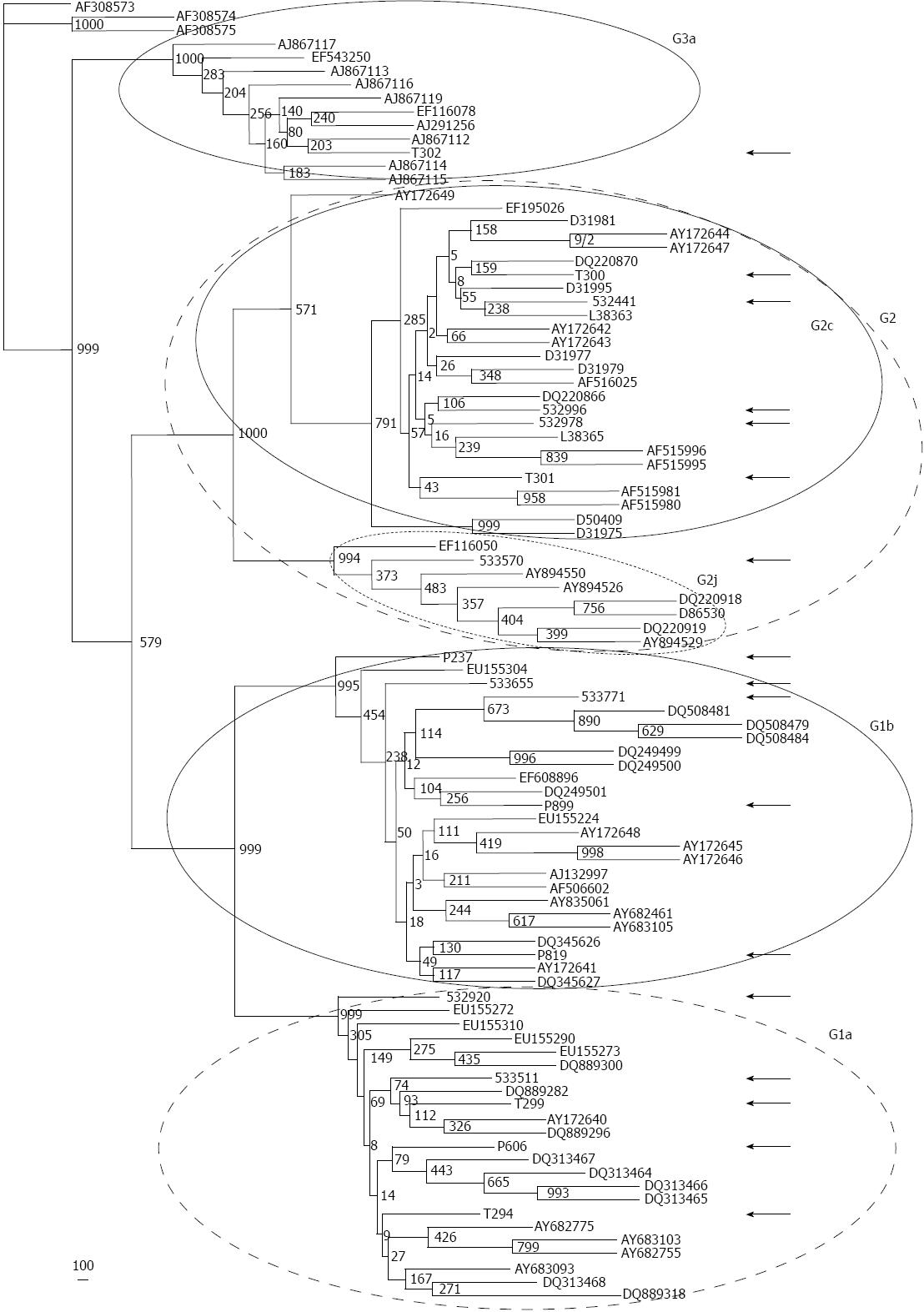
Figure 1 Assignment of Hepatitis C virus subtype isolates according to the phylogenetic tree performed by the Neighbor-Joining method, after an NS5B alignment of Hepatitis C virus sequences obtained from the 17 Argentinian volunteers studied herein, as well as from 89 reference sequences (e.
g., those composing subgenotype groups G1a, G1b, G2c, G2j, G3a) downloaded from GenBank. A consensus tree is shown after analyzing 1000 replicate trees. A distance scale (in nucleotide substitutions per position) is shown. Arrows indicate Hepatitis C virus sequences derived from Argentinian volunteers.
- Citation: del Pino N, Oubiña JR, Rodríguez-Frías F, Esteban JI, Buti M, Otero T, Gregori J, García-Cehic D, Camos S, Cubero M, Casillas R, Guàrdia J, Esteban R, Quer J. Molecular epidemiology and putative origin of hepatitis C virus in random volunteers from Argentina. World J Gastroenterol 2013; 19(35): 5813-5827
- URL: https://www.wjgnet.com/1007-9327/full/v19/i35/5813.htm
- DOI: https://dx.doi.org/10.3748/wjg.v19.i35.5813