Published online Dec 28, 2012. doi: 10.3748/wjg.v18.i48.7251
Revised: October 31, 2012
Accepted: November 11, 2012
Published online: December 28, 2012
AIM: To investigate the relationship between c.343A>G and c.2216A>C polymorphism sites in the CDH17 gene and colorectal carcinoma.
METHODS: Ninety-three non-consanguineous colorectal carcinoma patients admitted to the Department of Oncology at the First Affiliated Hospital of Zhengzhou University were included in this study. Ninety-three peripheral venous blood samples, of approximately one milliliter from each patient, were collected between December 2009 and August 2010. The genomic DNA of these peripheral venous blood samples were extracted and purified using a Fermentas Genomic DNA Purification Kit (Fermentas, CA) according to the manufacturer’s protocol. The single nucleotide polymorphisms (SNPs) of the liver-intestine cadherin (CDH17) gene c.343A>G and c.2216A>C were determined by the polymerase chain reaction-single strand conformation polymorphism method (PCR-SSCP) in 93 peripheral venous blood samples from patients suffering with colorectal carcinoma. Typical samples that showed different migration bands in SSCP were confirmed by sequencing. Directed DNA sequencing was used to check the correctness of the genotype results from the PCR-SSCP method.
RESULTS: There was a significant association between the c.2216 A>C SNPs of the CDH17 gene and the tumor-node-metastasis (TNM) grade, as well as with lymph node status, in 93 peripheral venous blood samples from colorectal carcinoma patients. The genotype frequencies of A/C, A/A, and C/C were 12.90%, 33.33% and 53.76%, respectively. There was a significant correlation between lymph node metastasis, TNM grade, and the genotype distribution (P < 0.05). The C/C genotype raised the risk of lymph node metastasis and the TNM grade. There was a significant difference in the TNM grade and lymph node metastasis between the A/A and C/C genotypes (P = 0.003 and P = 0.013, respectively). Patients with colorectal carcinoma carrying the C allele tended to have a higher risk of lymph node metastasis and have a higher TNM grade. The difference between the TNM grades, as well as the lymph node metastasis of the two alleles, was statistically significant (P < 0.01).
CONCLUSION: The SNPs of the CDH17 gene c.2216 A>C might be clinically important in the prognosis of colorectal carcinoma.
-
Citation: Chen RY, Cao JJ, Chen J, Yang JP, Liu XB, Zhao GQ, Zhang YF. Single nucleotide polymorphisms in the
CDH17 gene of colorectal carcinoma. World J Gastroenterol 2012; 18(48): 7251-7261 - URL: https://www.wjgnet.com/1007-9327/full/v18/i48/7251.htm
- DOI: https://dx.doi.org/10.3748/wjg.v18.i48.7251
Colorectal carcinoma (CRC) has traditionally been one of the four cancers with the highest mortality worldwide, but the incidence was relatively low historically in Asian populations. However, the prevalence of colorectal neoplasms has increased by 2- to 4-fold in some developed Asian countries, including China, Japan, South Korea and Singapore in the past few decades, due to changes in dietary habits and lifestyle, as well as certain genetic factors of Asian populations[1]. The etiology of CRC is multifactor, involving hereditary causes, environmental factors, and somatogenic changes occurring during tumor progression. At present, the assured mechanism of the occurrence and development of colorectal carcinoma is still unknown. While several aspects of colorectal carcinoma have been investigated, CRCs can be parsimoniously subdivided into two major groups defined by the genetic pathways involved[2]. Its correlation with single nucleotide polymorphisms (SNPs) remains largely unknown.
SNPs are the most common type of genomic sequence variation[3], and are thought to be associated with population diversity, susceptibility to disease, and individual response to drug treatments. Many SNPs are silent, with no direct effect on the gene products but, by virtue of the linkage disequilibrium existing across the human genome, they can still be used as genetic markers to locate adjacent functional variants that contribute to diseases. Foreseeable common SNP discoveries may not permit the identification of the small subset of patients that contain most cancers[4]. SNPs may also have functional consequences if they directly affect the coding or regulatory (usually promoter) regions of a gene. There have been cumulative studies on the associations between cancer risk and SNPs in selected candidate genes; to date, numerous SNPs associated with susceptibility to both cancer types have been identified, but their effect on disease risk may differ among populations[5], and such information may shed light on the molecular and genetic basis of the polygenic nature of cancer. These SNPs may be used as surrogate biomarkers of the genetic background of CRC patients, to predict therapeutic response and prognosis[6].
The CDH17 gene is a member of the cadherin superfamily; genes encoding calcium-dependent, membrane-associated glycoproteins. The human CDH17 gene located on chromosome 8q22.1 has eighteen exons and encodes liver-intestine cadherin (LI-cadherin) protein (also known as cadherin-17). LI-cadherin is a structurally unique member of the cadherin superfamily[7]. In contrast to classic cadherins, such as E- and N-cadherin, the extracellular domain of LI-cadherin consists of seven, instead of the usual five, structurally defined cadherin repeats. Its cytoplasmic domain is also small, comprising only 20 amino acids without a β-catenin binding region, and exhibits no homology with the corresponding region of classic cadherins, which consists of 150-160 amino acids. LI-cadherin is also known to possess biological functions distinct from classic cadherins. The adhesive function of LI-cadherin is independent of any interaction with cytoplasmic components, such as catenins or the actin cytoskeleton, although in E-cadherin this function crucially depends on the formation of a cadherin-catenin complex and anchorage to the actin cytoskeleton[8,9]. The mechanism responsible for the regulation and function of this cadherin has been elucidated. Expression of LI-cadherin has been reported in gastric adenocarcinoma of the intestinal type and colorectal carcinoma[10,11]. In colorectal carcinoma, LI-cadherin expression has been reported in well-differentiated carcinoma, but not in poorly-differentiated carcinoma. It has been found that reduced expression of LI-cadherin is closely associated with tumor progression and lymph node metastasis of human colorectal carcinoma[12]. However, the detailed clinicopathologic significance of LI-cadherin has not been elucidated.
In this study, through a PubMed, Embase, Google Scholar, CBMdisc and CNKI SNPs search, we found that for the CDH17 gene c.343A>G and c.2216A>C, the SNPs gene phenotype is A/G and the gene frequency phenotype is about 50%. We detected the SNPs of the CDH17 gene c.343A>G and c.2216A>C by polymerase chain reaction-single strand conformation polymorphism method (PCR-SSCP), and directed sequencing in colorectal carcinoma patients. We then analyzed the distribution frequencies of the genotypes and alleles of the two polymorphism sites. To evaluate the role of CDH17 gene polymorphisms in colorectal tumor aggressiveness, the associations between genotype and clinicopathologic parameters were also analyzed.
Ninety-three patients undergoing curative surgery for colorectal cancer at the First Affiliated Hospital of Zhengzhou University were included in this study after giving their informed consent. Peripheral blood samples were collected in ethylenediaminetetraacetic acid-containing tubes between December 2009 and August 2010 from these patients. All samples were immediately placed in refrigeratory conditions and stored at 4 °C until they could be further processed.
Genomic DNA was extracted using a Fermentas Genomic DNA Purification Kit (Fermentas, CA) according to the manufacturer’s protocol. The amount of isolated DNA was determined spectrophotometrically.
Primer sequences for the CDH17 gene c.343A>G and c.2216A>C were synthesized by Shanghai Sangon Biological Engineering Technology and Services Co. Ltd (Table 1).
| Gene | SNPs site (refSNP ) | Primer sequence(5’-3’) | Fragment sizes of PCR products (bp) |
| CDH17 | c.343A>G (rs2243518) | Forward primer: CCAACATGGTTTCCTTTTCCTC | 159 |
| Reverse primer: GTTCTGCCTTACTGAGCCTTCG | |||
| c.2216A>C (rs1051624) | Forward primer: AATCCAGGGTCTGAAGTTGTA | 198 | |
| Reverse primer: TACTAGCCTGAGTTGCCTATA |
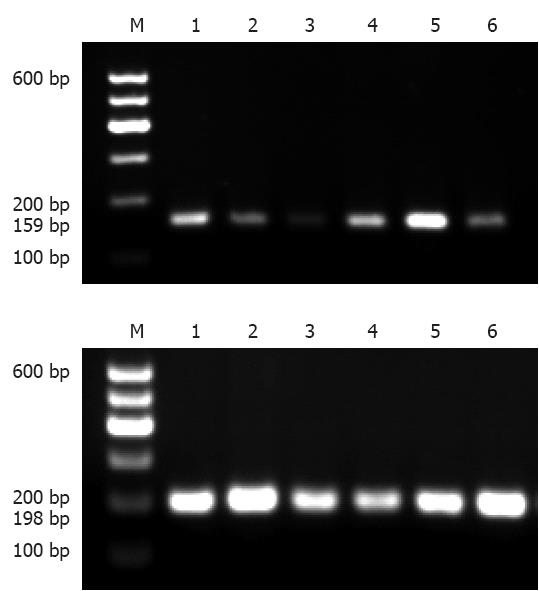
The fragments encompassing the CDH17 gene c.343A>G and c.2216A>C were amplified by PCR. The twenty microlitre PCR amplification reaction system contained < 1 μg template DNA, 10 μL of 2 × Taq PCR Master Mix [Tiangen Biotech (Beijing) Co., Ltd.], 0.4 μL of forward and reverse primers (10 μmol/L), and 3.2 μL of double distilled water. PCR reaction conditions were as follows: initial denaturation at 94 °C for 5 min, followed by 35 cycles of denaturation at 94 °C for 30 s, annealing for 30 s and extension at 72 °C for 1 min, and a final extension at 72 °C for 10 min, with preservation being carried out at 4 °C. After 2.0% agarose gel electrophoresis (containing Evans blue dye), PCR products were observed by a gel imaging instrument. Fragment sizes of PCR products were 159 bp and 198 bp, respectively, and indicated with DL100 DNA Marker.
The amplified products were denatured in 95% formamide at 95 °C for 10 min, and analyzed on 12% polyacrylamide gels at 160 V for 15 min and 30 V for 6-8 h in 1 × Tris-Borate-EDTA buffer at normal body temperature. A modified silver staining method was applied for visualization. Briefly, the gels were soaked in 10% ethanol and 0.5% acetic acid for 10 min, then transferred to 0.2% AgNO3 solution for 10 min and washed with distilled water three times. The staining reaction was developed with the use of 3% NaOH and 0.25% formaldehyde, and the gels were fixed in 10% ethanol and 0.5% acetic acid. Staining reactions were promoted by gentle shaking.
The detection of SSCP analysis was performed. Samples showing an altered mobility pattern were then subjected to sequencing analysis by Shanghai Sangon Biological Engineering Technology and Services Co. Ltd.
Statistical analysis was performed using SPSS for Windows version 17.0. Differences in allele and genotype frequencies were evaluated by the χ2 or Fisher exact test. We used logistic regression models to calculate χ2 tests for genotypic and allelic association and odds ratios (OR) with their 95%CI. Association between each of the SNPs and clinicopathological parameters was assessed using the χ2 test. To study correlation between clinical parameters and genotypes of the CDH17 gene c.343A>G and c.2216A>C polymorphisms in multivariate mode we used binary logistic regression, with each parameter as a dependent variable, and genotypes of the two studied SNPs together with other clinical parameters as independent variables. Differences in ordered categorical data were evaluated for significance with the rank sum test; the mean ranks were used to express risk degree. The Holm-Bonferroni method was used for multiple comparisons. A P value of ≤ 0.05 was considered statistically significant.
The characteristics of 93 patients with colorectal carcinoma are given in Tables 2 and 3. Of the 93 patients, 49% (46/93) were female and 51% (47/93) were male. The mean age of the female patients (the mean ± SD deviation was 56.41 ± 14.91 years) did not differ significantly from that of the males (the mean ± SD deviation was 58.19 ± 11.60 years).
| Clinicopathologic parameters | n | Genotype CDH17 c.343A>G | χ2 | P value | ||
| A/A | A/G | G/G | ||||
| Gender | ||||||
| Male | 47 | 10 (21.3) | 25 (53.2) | 12 (25.5) | 2.236 | 0.327 |
| Female | 46 | 16 (34.8) | 19 (41.3) | 11 (23.9) | ||
| Age (yr) | ||||||
| < 50 | 29 | 10 (34.5) | 15 (51.7) | 4 (13.8) | 2.854 | 0.240 |
| ≥ 50 | 64 | 16 (25) | 29 (45.3) | 19 (29.7) | ||
| Tumor location | ||||||
| Colon | 32 | 8 (25.0) | 16 (50.0) | 8 (25.0) | 0.229 | 0.892 |
| Rectum | 61 | 18 (29.5) | 28 (45.9) | 15 (24.6) | ||
| Tumor size (cm) | ||||||
| < 5 | 68 | 18 (26.5) | 34 (50.0) | 16 (23.5) | 0.734 | 0.693 |
| ≥ 5 | 25 | 8 (32.0) | 10 (40.0) | 7 (28.0) | ||
| Macroscopic appearance | ||||||
| Protrude type | 31 | 8 (25.8) | 17 (54.8) | 6 (19.4) | 1.948 | 0.7451 |
| Ulcer type | 46 | 12 (26.1) | 21 (45.7) | 13 (28.3) | ||
| Infiltrating type | 16 | 6 (37.5) | 6 (37.5) | 4 (25.0) | ||
| Colloid type | 0 | 0 (0.0) | 0 (0.0) | 0 (0.0) | ||
| Histologic type | ||||||
| Adenocarcinoma | 71 | 18 (25.4) | 34 (47.9) | 19 (26.8) | 2.179 | 0.7031 |
| Adenocarcinoma and mucinous carcinoma | 9 | 4 (44.4) | 3 (33.3) | 2 (22.2) | ||
| Mucinous carcinoma | 13 | 4 (30.8) | 7 (53.8) | 2 (15.4) | ||
| Undifferentiated carcinoma | 0 | 0 (0.0) | 0 (0.0) | 0 (0.0) | ||
| Adenosquamous carcinoma | 0 | 0 (0.0) | 0 (0.0) | 0 (0.0) | ||
| Squamous cell carcinoma | 0 | 0 (0.0) | 0 (0.0) | 0 (0.0) | ||
| Invasion depth | ||||||
| Submucosa | 2 | 0 (0.0) | 2 (100.0) | 0 (0.0) | 0.696 | 0.7062 |
| Muscular layer | 34 | 12 (35.3) | 15 (44.1) | 7 (20.6) | ||
| Serous coat | 48 | 12 (25) | 21 (43.8) | 15 (31.3) | ||
| Other organs | 9 | 2 (22.2) | 6 (66.7) | 1 (11.1) | ||
| Differentiation degree | ||||||
| Well-differentiated | 4 | 0 (0.0) | 2 (50.0) | 2 (50.0) | 2.212 | 0.3312 |
| Moderately-differentiated | 63 | 17 (27.0) | 33 (52.4) | 13 (20.6) | ||
| Poorly-differentiated | 26 | 9 (34.6) | 9 (34.6) | 8 (30.8) | ||
| Lymph node metastasis | ||||||
| No | 59 | 16 (27.1) | 35 (59.3) | 8 (13.6) | 13.105 | 0.0013 |
| Yes | 34 | 10 (29.4) | 9 (26.5) | 15 (44.1) | ||
| TNM grade | ||||||
| I | 26 | 8 (30.8) | 16 (61.5) | 2 (7.7) | 10.344 | 0.00623 |
| II | 31 | 7 (22.6) | 18 (58.1) | 6 (19.4) | ||
| III | 33 | 9 (27.3) | 9 (27.3) | 15 (45.5) | ||
| IV | 3 | 2 (66.7) | 1 (33.3) | 0 (0.0) | ||
| Hematogenous metastasis | ||||||
| No | 90 | 24 (26.7) | 43 (47.8) | 23 (25.6) | 2.556 | 0.2791 |
| Yes | 3 | 2 (66.7) | 1 (33.3) | 0 (0.0) | ||
| Recurrence | ||||||
| No | 91 | 24 (26.4) | 44 (48.4) | 23 (25.3) | 5.267 | 0.0721 |
| Yes | 2 | 2 (100.0) | 0 (0.0) | 0 (0.0) | ||
| Clinicopathologic parameters | n | Genotype CDH17 c.2216A>G | χ2 | P value | ||
| A/A | A/C | C/C | ||||
| Gender | ||||||
| Male | 47 | 20 (42.6) | 5 (10.6) | 22 (46.8) | 3.656 | 0.161 |
| Female | 46 | 11 (23.9) | 7 (15.2) | 28 (60.9) | ||
| Age (yr) | ||||||
| < 50 | 29 | 6 (20.7) | 5 (17.2) | 18 (62.1) | 3.176 | 0.204 |
| ≥ 50 | 64 | 25 (39.1) | 7 (10.9) | 32 (50.0) | ||
| Tumor location | ||||||
| Colon | 32 | 10 (31.3) | 6 (18.8) | 16 (50.0) | 1.485 | 0.476 |
| Rectum | 61 | 21 (34.4) | 6 (9.8) | 34 (55.7) | ||
| Tumor size (cm) | ||||||
| < 5 | 68 | 25 (36.8) | 5 (7.4) | 38 (55.9) | 7.144 | 0.0283 |
| ≥ 5 | 25 | 6 (24.0) | 7 (28.0) | 12 (48.0) | ||
| Macroscopic appearance | ||||||
| Protrude type | 31 | 11 (35.5) | 4 (12.9) | 16 (51.6) | 1.025 | 0.9061 |
| Ulcer type | 46 | 16 (34.8) | 5 (10.9) | 25 (54.3) | ||
| Infiltrating type | 16 | 4 (25.0) | 3 (18.8) | 9 (56.3) | ||
| Colloid type | 0 | 0 (0.0) | 0 (0.0) | 0 (0.0) | ||
| Histologic type | ||||||
| Adenocarcinoma | 71 | 23 (32.4) | 10 (14.1) | 38 (53.5) | 1.684 | 0.7941 |
| Adenocarcinoma and mucinous carcinoma | 9 | 4 (44.4) | 0 (0.0) | 5 (55.6) | ||
| Mucinous carcinoma | 13 | 4 (30.8) | 2 (15.4) | 7 (53.8) | ||
| Undifferentiated carcinoma | 0 | 0 (0.0) | 0 (0.0) | 0 (0.0) | ||
| Adenosquamous carcinoma | 0 | 0 (0.0) | 0 (0.0) | 0 (0.0) | ||
| Squamous cell carcinoma | 0 | 0 (0.0) | 0 (0.0) | 0 (0.0) | ||
| Invasion depth | ||||||
| Submucosa | 2 | 2 (100.0) | 0 (0.0) | 0 (0.0) | 1.610 | 0.4472 |
| Muscular layer | 34 | 12 (35.3) | 4 (11.8) | 18 (52.9) | ||
| Serous coat | 48 | 14 (29.2) | 5 (10.4) | 29 (60.4) | ||
| Other organs | 9 | 3 (33.3) | 3 (33.3) | 3 (33.3) | ||
| Differentiation degree | ||||||
| Well-differentiated | 4 | 0 (0.0) | 0 (0.0) | 4 (100.0) | 0.090 | 0.9562 |
| Moderately-differentiated | 63 | 24 (38.1) | 9 (14.3) | 30 (47.6) | ||
| Poorly-differentiated | 26 | 7 (26.9) | 3 (11.5) | 16 (61.5) | ||
| Lymph node metastasis | ||||||
| No | 59 | 25 (42.4) | 10 (16.9) | 24 (40.7) | 11.143 | 0.0043 |
| Yes | 34 | 6 (17.6) | 2 (5.9) | 26 (76.5) | ||
| TNM grade | ||||||
| I | 26 | 12 (46.2) | 4 (15.4) | 10 (38.5) | 6.710 | 0.03523 |
| II | 31 | 12 (38.7) | 5 (16.1) | 14 (45.2) | ||
| III | 33 | 6 (18.2) | 2 (6.1) | 25 (75.8) | ||
| IV | 3 | 1 (33.3) | 1 (33.3) | 1 (33.3) | ||
| Hematogenous metastasis | ||||||
| No | 90 | 30 (33.3) | 11 (12.2) | 49 (54.4) | 1.243 | 0.5371 |
| Yes | 3 | 1 (33.3) | 1 (33.3) | 1 (33.3) | ||
| Recurrence | ||||||
| No | 91 | 29 (31.9) | 12 (13.2) | 50 (54.9) | 4.088 | 0.1301 |
| Yes | 2 | 2 (100.0) | 0 (0.0) | 0 (0.0) | ||
The PCR-SSCP analysis generated two fragments as expected; 159bp for c.343A>G, and 198bp for c.2216A>C (Figure 1).
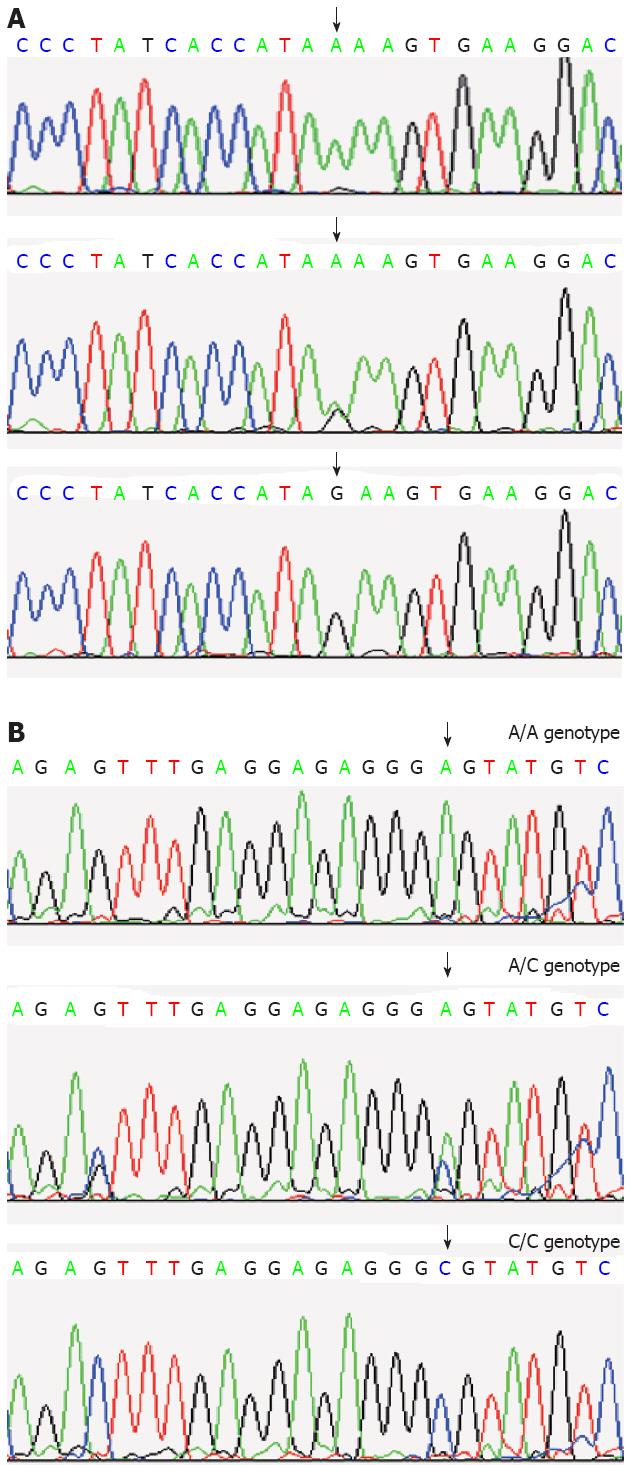
The genotype of the CDH17 gene c.343A>G: The genotype frequencies of the CDH17 gene c.343A>G A/A, A/G, and G/G were 27.96% (26/93), 47.31% (44/93), and 24.73% (23/93), respectively. The frequencies of the A allele and G allele were 51.61% and 48.39%, respectively.
The correlation between the genotype of the CDH17 gene c.343A>G and clinicopathologic parameters: There were no significant differences in gender, age, tumor location, tumor size, macroscopic appearance, histologic type, invasion depth, differentiation, hematogenous metastasis, or recurrence among the three genotypes of the CDH17 gene c.343A>G (P > 0.05). However, there was a significant correlation between lymph node metastasis and tumor-node-metastasis (TNM) grade, and its genotype frequencies (P < 0.05) (Table 1).
The sequencing analysis: The detection of SSCP analysis was performed. Samples showing an altered mobility pattern were then subjected to sequencing analysis by Shanghai Sangon Biological Engineering Technology and Services Co. Ltd (Figure 2).
The correlation between the genotype of the CDH17 gene c.343A>G and lymph node metastasis: The results of logistic regression analysis are shown in Table 4, and indicate that the G/G genotype of the CDH17 gene c.343A>G raised the risk of lymph node metastasis (OR of genotype G/G, genotype A/A to genotype A/G were 7.292 and 2.431, respectively). The results of multiple comparisons by the Holm-Bonferroni method (α’ = 0.017) were as follows: P value was 0.102 when genotype A/A was compared with genotype A/G; P value was 0.062 when genotype A/A was compared with genotype G/G; P value was 0.000 when genotype A/G was compared with genotype G/G. This indicates that there was a significant difference in lymph node metastasis between the A/G and G/G genotypes.
| Lymph node metastasis | OR (95%CI) | ||
| No | Yes | ||
| Genotype | |||
| A/G | 35 (59.3) | 9 (26.5) | 1.000 (reference) |
| A/A | 16 (27.1) | 10 (29.4) | 2.431 (0.828-7.139) |
| G/G | 8 (13.6) | 15 (44.1) | 7.292 (2.360-22.532) |
| Allele frequency | |||
| A | 67 (56.8) | 29 (42.6) | 1.000 (reference) |
| G | 51 (43.2) | 39 (57.4) | 1.767 (0.967-3.229) |
The CDH17 gene c.343A>G site allele G raised the risk of lymph node metastasis (OR of allele G to allele A was 1.767) (Table 4). However, a χ2 test showed that the difference between the lymph node metastasis of the two alleles was not statistically significant (χ2 = 3.450; df = 1; P = 0.063).
The correlation between the genotype of the CDH17 gene c.343A>G and TNM grade: The results of the Kruskal-Wallis test are shown in Table 5. It revealed that the CDH17 gene c.343A>G genotype G/G raised the risk of TNM grade (the mean ranks of A/G, A/A and G/G genotype were 39.32, 48.15 and 60.39, respectively). The results of multiple comparisons by the Holm-Bonferroni method (α’ = 0.017) were as follows: P value was 0.197 when genotype A/A was compared with genotype A/G; P value was 0.127 when genotype A/A was compared with genotype G/G; P value was 0.001 when genotype A/G was compared with genotype G/G. This suggests that there was a significant difference in TNM grade between the A/G and G/G genotypes.
| TNM grade | Mean rank | ||||
| I | II | III | IV | ||
| Genotype | |||||
| A/G | 16 (61.5) | 18 (58.1) | 9 (27.3) | 1 (33.3) | 39.32 |
| A/A | 8 (30.8) | 7 (22.6) | 9 (27.3) | 2 (66.7) | 48.15 |
| G/G | 2 (7.7) | 6 (19.4) | 15 (45.5) | 0 (0.0) | 60.39 |
| Allele frequency | |||||
| A | 32 (61.5) | 32 (51.6) | 27 (40.9) | 5 (83.3) | 87.71 |
| G | 20 (38.5) | 30 (48.4) | 39 (59.1) | 1 (16.7) | 99.68 |
The genotype of the CDH17 gene c.2216A>C: The genotype frequencies of the CDH17 gene c.2216A>C A/C, A/A, and C/C were 12.90% (12/93), 33.33% (31/93), and 53.76% (50/93), respectively. The frequencies of the A allele and C allele were 39.78% and 60.22%, respectively.
The allele G raised the risk of TNM grade (the mean ranks of allele A and allele G were 87.71 and 99.68, respectively) (Table 5). However, the Kruskal-Wallis test showed that the difference between the TNM grade of the two alleles was not statistically significant (H = 2.561; df = 1; P = 0.110).
The correlation between the genotype of the CDH17 gene c.2216A>C and clinicopathologic parameters: There was no significant difference in gender, age, tumor location, macroscopic appearance, histologic type, invasion depth, differentiation, hematogenous metastasis, or recurrence among the three genotypes of the CDH17 gene c.2216A>C (P > 0.05). However, there was a significant correlation between tumor size, lymph node metastasis, and TNM grade, and its genotype frequencies (P < 0.05) (Table 3).
The correlation between the genotype of the CDH17 gene c.2216A>C and tumor size: The results of the χ2 test indicate that there was a significant correlation between tumor size and the genotype of the CDH17 gene c.2216A>C. However, the results of multiple comparisons by the Holm-Bonferroni method (α’ = 0.017) suggest that there was no significant difference in tumor size between any of its genotypes (P value was 0.018 when genotype A/A was compared with genotype A/C; P value was 0.625 when genotype A/A was compared with genotype C/C; P value was 0.027 when genotype A/C was compared with genotype C/C).
The correlation between the genotype of the CDH17 gene c.2216A>C and lymph node metastasis: The results of logistic regression analysis are shown in Table 6, and indicate that the C/C genotype of the CDH17 gene c.2216A>C raised the risk of lymph node metastasis (OR of genotype C/C, genotype A/A to genotype A/C were 5.417 and 1.200, respectively). The results of multiple comparisons by the Holm-Bonferroni method (α’ = 0.017) were as follows: P value was 0.067 when genotype A/A was compared with genotype A/C; P value was 0.003 when genotype A/A was compared with genotype C/C; P value was 0.027 when genotype A/C was compared with genotype C/C. This indicates that there was a significant difference in lymph node metastasis between the A/A and C/C genotypes.
| Lymph node metastasis | OR (95%CI) | ||
| No | Yes | ||
| Genotype | |||
| A/C | 10 (16.9) | 2 (5.9) | 1.000 (reference) |
| A/A | 25 (42.4) | 6 (17.6) | 1.200 (0.206-6.977) |
| C/C | 24 (40.7) | 26 (76.5) | 5.417 (1.076-27.272) |
| Allele frequency | |||
| A | 60 (50.8) | 14 (20.6) | 1.000 (reference) |
| C | 58 (49.2) | 54 (79.4) | 3.990 (2.002-7.953) |
CDH17 gene c.2216A>C site allele C raised the risk of lymph node metastasis (OR of allele C to allele A was 3.990) (Table 6). The χ2 test showed that the difference between the lymph node metastasis of the two alleles was statistically significant (χ2 = 16.488; df = 1; P = 0.000).
The correlation between the genotype of the CDH17 gene c.2216A>C and TNM grade: The results of the Kruskal-Wallis test are shown in Table 7, and reveal that the CDH17 gene c.2216A>C genotype C/C raised the risk of TNM grade (the mean ranks of the A/A, A/C, and C/C genotype were 38.77, 42.00 and 53.30, respectively). The results of multiple comparisons by the Holm-Bonferroni method (α’ = 0.017) were as follows: P value was 0.718 when genotype A/A was compared with genotype A/C; P value was 0.013 when genotype A/A was compared with genotype C/C; P value was 0.168 when genotype A/C was compared with genotype C/C. This suggests that there was a significant difference in TNM grade between the A/A and C/C genotypes.
| TNM grade | Mean rank | ||||
| I | II | III | IV | ||
| Genotype | |||||
| A/C | 4 (15.4) | 5 (16.1) | 2 (6.1) | 1 (33.3) | 42 |
| A/A | 12 (46.2) | 12 (38.7) | 6 (18.2) | 1 (33.3) | 38.77 |
| C/C | 10 (38.5) | 14 (45.2) | 25 (75.8) | 1 (33.3) | 53.3 |
| Allele frequency | |||||
| A | 28 (53.8) | 29 (46.8) | 14 (21.2) | 3 (50.0) | 78.09 |
| C | 24 (46.2) | 33 (53.2) | 52 (78.8) | 3 (50.0) | 103.68 |
The Kruskal-Wallis test showed that allele C raised the risk of TNM grade (the mean ranks of allele A and allele C were 78.09 and 103.68, respectively) (Table 7). The difference between the TNM grade of the two alleles was statistically significant (H = 11.225; df = 1; P = 0.001).
Cancer is a complex disease where genetic mutations, rearrangement, deletions, or gene polymorphisms may affect not only cancer development, but also cancer progression and, as a result, could influence cancer phenotypes[13,14]. The CDH17 gene is located on chromosome 8q22.1 and encodes LI-cadherin. LI-cadherin, belonging to a subclass of the 7D-cadherin superfamily, is present in the fetal liver and gastrointestinal tract during embryogenesis, but the gene becomes silenced in healthy adult liver and stomach tissues. It functions as a peptide transporter and cell adhesion molecule to maintain tissue integrity in epithelia. However, recent findings have reported aberrant expression of LI-cadherin in major gastrointestinal malignancies, including hepatocellular carcinoma (HCC), stomach and colorectal cancers, and its clinical association with tumor metastasis and advanced tumor stages[15,16]. LI-cadherin manifests distinct and unique roles in tumorigenesis originating from different organs. In the epithelium of the healthy human stomach, the level of LI-cadherin expression is negligible, whereas an overexpression of this molecule is observed frequently in gastric carcinoma[17-19]. When the transcriptome of gastric cancer was studied by serial analysis of gene expression, LI-cadherin was found associating with an intestinal type of gastric cancer[20]. In addition, high tumoral LI-cadherin levels tend to correlate with advanced stages of gastric cancer, and are associated with a poor prognosis and lymph node metastasis[21,22]. That silencing Li-cadherin has positive actions in the processes of LoVo cell invasion and metastasis, and the interactions among matrix metalloproteinase (MMP)-2, MMP-9, and Li-cadherin, participates in the multiple steps of invasion and metastasis in LoVo colorectal cancer cells[23].
This observation was further supported by a study using high density DNA microarrays to identify differentially-expressed genes in advanced stages of gastric cancer, in which an upregulation of CDH17 was found among genes associated with cell adhesion, cell cycle, cellular motility, and DNA synthesis[24]. Unlike normal gastric tissues, LI-cadherin is found scattered throughout the healthy human pancreas. Less LI-cadherin is expressed in poorly-differentiated tumor tissues of ductal adenocarcinoma of the pancreas, and this low level of LI-cadherin is correlated with advanced stages of the cancer. When LI-cadherin is used as a prognostic biomarker, the survival time is longer for those patients with LI-cadherin-positive tumors when compared with those having LI-cadherin-negative tumors[25]. In the midst of identifying new target for HCC detection and treatment, LI-cadherin is likely an attractive candidate[26]. The elevated level of LI-cadherin in tumors is correlated to high serum alpha-fetoprotein level, tumor invasion, and advanced stage tumor, associating with poor prognosis, of HCC patients. For primary colorectal carcinoma, the expression of LI-cadherin is diminished in tumor tissues. Clinically, this low level of LI-cadherin is correlated to dedifferentiation of tumors, tumor invasion, and late tumor stages. However, significant differences in the survival of cancer patients have not been found, irrespective of whether the expressions of LI-cadherin are low or high. In addition, a negligible level of LI-cadherin has been documented in several colorectal cancer cell lines when compared with those in normal colonic mucosal cells[27]. This observation is further supported by a separate study showing a reduced expression of LI-cadherin in cancer tissues and such expression was correlated with dedifferentiation of tumors and poor survival of patients[28]. But Gröne et al[29] study showed CDH17 protein and gene expression analyses do not support application of molecular classifiers for prediction of clinical outcome in current routine diagnostic as a basis for patient-orientated therapy in stage Union for International Cancer Control II colon cancer. Further studies are needed to develop prognosis signatures applicable in patient care. Despite these findings, LI-cadherin was found present in colorectal adenocarcinoma in a separate study[30].
In this study we investigated the SNPs in c.343A>C and c.2216A>C polymorphism sites of CDH17 gene and the associations between genotype of them and clinicopathologic parameters were also analyzed to evaluate the role of SNPs in colorectal tumor aggressiveness. The possibility that different genetic polymorphisms in the CDH17 gene may regulate, at least in part, LI-cadherin expression and/or activity, is an attractive hypothesis that may help to identify patients who are susceptible to colorectal carcinoma.
There have been many studies on the associations between cancer risk and SNPs. But the associations between CDH17 gene SNPs in c.343A>C and c.2216A>C polymorphism sites and colorectal carcinoma risk have not been reported.
In our study, the first missense substitution (c.343A>G) mapped to the amino acid position 115 and was responsible for the amino acid change of 115 Glu with Lys. The genotype frequencies of A/A, A/G and G/G were 27.96%, 47.31% and 24.73%, respectively. The frequencies of the A allele and G allele were 51.61% and 48.39%, respectively. According to the SNP database (rs2243518), the frequency of the G allele in our population is lower than that in Europeans (84.2%). In comparison, its reported frequency in Sub-Saharan African and Asians is 95.8% and 56.7%, respectively. There were no significant difference in gender, age, tumor location, tumor size, macroscopic appearance, histologic type, invasion depth, differentiation, hematogenous metastasis, and recurrence among the genotype distribution (P > 0.05). But we found a significant correlation between the genotype distribution and lymph node metastasis and TNM grade (P < 0.05). The results of logistic regression analysis indicate that the G/G genotype raised risk of lymph node metastasis (OR of genotype G/G, genotype A/A to genotype A/G were 7.292 and 2.431, respectively). The results of multiple comparison by Holm-Bonferroni (α’ = 0.017) indicate that there was a significant difference in lymph node metastasis between the A/G and G/G genotypes (P = 0.000). And patients with colorectal carcinoma carrying the G allele tended to have a higher risk of lymph node metastasis (OR of allele G to allele A was 1.767). But χ2 test showed that the difference between the lymph node metastasis of two alleles was not statistically significant (χ2 = 3.450; df = 1; P = 0.063). In addition, the results of Kruskal-Wallis revealed that the genotype G/G raised risk of TNM grade (the mean ranks of A/G, A/A and G/G genotype were 39.32, 48.15 and 60.39, respectively). And the results of multiple comparison by Bonferroni (α’ = 0.017) suggest that there was a significant difference in TNM grade between the A/G and G/G genotypes (P = 0.001). Patients with colorectal carcinoma carrying the G allele tended to have a higher TNM grade (the mean ranks of allele A and allele G were 87.71 and 99.68, respectively). But the Kruskal-Wallis test showed that the difference between the TNM grade of two alleles was not statistically significant (H = 2.561; df = 1; P = 0.110).
The second SNP we investigate here (c.2216A>C) mapped to the amino acid position 739 and was responsible for the amino acid change of 739 Ala with Glu. The genotype frequencies of A/C, A/A and C/C were 12.90%, 33.33% and 53.76%, respectively. The frequencies of the A allele and C allele were 39.78% and 60.22%, respectively. According to the SNP database (rs1051624), the frequency of the C allele in our population is higher than that in Europeans (58.3%). In comparison, its reported frequency in African-American and Asians is 23.9% and 58.3%, respectively. There were no significant difference in gender, age, tumor location, macroscopic appearance, histologic type, invasion depth, differentiation, hematogenous metastasis, and recurrence among the genotype distribution (P > 0.05). But there were a significant correlation between tumor size, lymph node metastasis and TNM grade and the genotype distribution (P < 0.05). Firstly, the results of χ2 test indicate that there was a significant correlation between tumor size and the genotype distribution. But the results of multiple comparison by Holm-Bonferroni (α’ = 0.017) suggest that there were no significant difference in tumor size between any two genotypes (P > 0.017). But Kwak et al[31] study showed that reduced expression of liver intestine-cadherin had a significant correlation with tumoral dedifferentiation and short overall survival in this series. In addition, early and frequent loss of liver intestine-cadherin expression might be a more sensitive indicator than E-cadherin to predict more aggressive tumoral behavior. Secondly, the results of logistic regression analysis indicate that the C/C genotype raised risk of lymph node metastasis (OR of genotype C/C, genotype A/A to genotype A/C were 5.417 and 1.200, respectively). And the results of multiple comparison by Holm-Bonferroni (α’ = 0.017) indicate that there was a significant difference in lymph node metastasis between the A/A and C/C genotype (P = 0.003). And patients with colorectal carcinoma carrying the C allele tended to have a higher risk of lymph node metastasis (OR of allele C to allele A was 3.990). The χ2 test showed that the difference between the lymph node metastasis of two alleles was statistically significant (χ2 = 16.488; df = 1; P = 0.000). Lastly, the results of Kruskal-Wallis test revealed that genotype C/C raised risk of TNM grade (the mean ranks of A/A, A/C and C/C genotype were 38.77, 42.00 and 53.30, respectively). And the results of multiple comparison by Holm-Bonferroni (α’ = 0.017) suggest that there was a significant difference in TNM grade between the A/A and C/C genotypes (P = 0.013). Patients with colorectal carcinoma carrying the C allele tended to have a higher TNM grade (the mean ranks of allele A and allele C were 78.09 and 103.68, respectively). And the difference between the TNM grade of two alleles was statistically significant (H = 11.225; df = 1; P = 0.001). SNPs of the CDH17 gene c.2216A>C and lymph node metastasis deserve further consideration in future studies as a clinical indicator during presurgical evaluation. And we found here patients with colorectal carcinoma carrying the C allele tended to have a higher TNM grade. This suggests that this SNPs would be a good predictor of lymph node status and TNM grade prior to surgical intervention and thereby provides an indicator for deciding whether chemotherapy should be given or not.
It has been reported[32] that CDH17 mRNA is alternatively spliced to produce at least two variant transcripts, one of which was with exon 7 skipping and another with both exon 6 and exon 7 skipped. The consequences of both splicing patterns would introduce premature translational stop codon in the reading frame, thereby producing either nonfunctional protein or potentially dominant negative protein. And CDH17 aberrant splicing was highly associated with tumor dissemination and shorter survival of HCC patients. It has been showed that aberrant mRNA splicing of the CDH17 gene in liver tissues was triggered by the specific constellation of two CDH17 single nucleotide polymorphisms (651T and IVS6 + 35G). And the functional T-G haplotype of CDH17 (651C>T and IVS6 + 35A>G) is a genetic susceptibility factor for the development of HCC in a Chinese population[33]. CDH17 becomes an attractive target for HCC therapy[16]. And it has been proposed that LI-cadherin is a useful immunohistochemical marker for diagnosis of adenocarcinomas of the digestive system[25] and CDH17 may be an oncogene up-regulating invasive feature of gastric cancer cells and could be a hopeful target for the control of gastric cancer progression[34]. Moreover, it has been revealed that LI-cadherin participates in the multiple steps of invasion and metastasis in LoVo colorectal cancer cells[35,36]. In our study we investigated the SNPs in c.343A>C and c.2216A>C polymorphism sites of CDH17 gene. The c.343A>G polymorphism site mapped to the amino acid position 115 and was responsible for the amino acid change of 115 Glu with Lys. The c.2216A>C polymorphism site mapped to the amino acid position 739 and was responsible for the amino acid change of 739 Ala with Glu. We found that patients with colorectal carcinoma carrying the C allele of c.2216A>C tended to have a higher risk of lymph node metastasis and a higher TNM grade. The reason is possible that the SNPs of the c.2216A>C cause the amino acid change and result in the LI-cadherin expression and/or activity changes. The polymorphism of c.343A>G also causes the amino acid change. But there is no significant difference between c.343A>G SNPs and TNM grade as well as lymph node metastasis between the A and G alleles. It is possibly because that the CDH17 mRNA is spliced with the polymorphism site skipped (exon 6).
In summary, in a Chinese Han colorectal carcinoma patient population of Henan province, a significant association was found between c.2216A>C SNPs of CDH17 gene (rs1051624) and TNM grade as well as with lymph node status. With logistic regression analysis a correlation between TNM grades as well as with lymph node metastasis and C/C genotype of CDH17 gene c.2216A>C was found. And patients with colorectal carcinoma carrying the C allele tended to have a higher risk of lymph node metastasis and a higher TNM grade, suggesting that this polymorphism might be clinically important in prognosis. Validation and functional studies are needed to assess the significance of these associations in clinical practice.
We thank Yuan-Ting Gu for their wisdom and guidance in the retrieval and evaluation of these samples, and Shui-Jun Zhang, Shao-Min Zhang for assistance with acquisition of historical reference. This paper is written in tribute to professor Sugimura and with respect to other former faculty members and staff in the department of pathology at the First Affiliated Hospital of Zhengzhou University for the care and preservation of records and tissue samples from the pre-intensivist era. We particularly thank Dond-Ling Gao who worked on the technical aspects of tissue retrieval and preparation.
Colorectal carcinoma (CRC) has been the fourth place of common cancer incidence rate in the China. The etiology of CRC is multifactorial, involving hereditary causes, environmental factors, and somatogenic changes occurring during tumor progression. The single nucleotide polymorphisms (SNPs), as the third generation of heredity markers, are the most common type of genomic sequence variations, which are thought to be associated with population diversity, susceptibility to diseases, and individual response to drug treatments. It has become one of the hot spots in research. There have been cumulative studies on the associations between disease risk and SNPs in selected candidate genes.CDH17 gene, also known as cadherin-17 and liver-intestine cadherin (LI-cadherin), is a member of the cadherin superfamily, genes encoding calcium-dependent, membrane-associated glycoproteins. The human CDH17 gene located on chromosome 8q22.1 has eighteen exons and encodes LI-cadherin protein. There has been found that reduced expression of LI-cadherin protein is closely associated with tumor progression and lymph node metastasis of human colorectal carcinoma.
At present, the research of relation between CDH17 gene SNPs and diseases are relatively less at home and abroad. In this study, authors detected the genotype of the CDH17 gene c.343A>G and c.2216A>C SNPs sites by the polymerase chain reaction-single strand conformation polymorphism and directed DNA sequencing in ninety-three colorectal carcinoma patients. The objective of this study is to analyze the phenotype and distribution frequencies of genotype and allele of the CDH17 gene c.343A>G and c.2216A>C polymorphism sites and to evaluate the correlation between the two polymorphism sites of CDH17 gene and tumor occurrence and development as well as clinicopathologic parameters of colorectal carcinoma.
There were no significant difference in gender, age, tumor location, tumor size, macroscopic appearance, histologic type, invasion depth, differentiation, hematogenous metastasis, and recurrence among the genotypes of CDH17 gene c.343A>G. But there were a significant correlation between lymph node metastasis and Tumor-node-metastasis (TNM) grade and the genotype frequencies of it. Colorectal carcinoma patients carrying the G/G genotype have a higher risk of lymph node metastasis and a higher TNM grade compared with those carrying A/G genotype. There were no significant difference in gender, age, tumor location, tumor size, macroscopic appearance, histologic type, invasion depth, differentiation, hematogenous metastasis, and recurrence among the genotypes of CDH17 gene c.2216A>C. But there were a significant correlation between lymph node metastasis and TNM grade and the genotype frequencies of it. The C/C genotype and C allele of this SNPs site might be a risk factor for a higher risk of lymph node metastasis and a higher TNM grade of colorectal carcinoma patient.
The SNPs of CDH17 gene c.2216 A>C might be clinically important in prognosis of colorectal carcinoma.
The study analyzed polymorphisms of CDH17 gene in blood samples collected from 93 colorectal carcinoma patients and found a strong association between c.2216A>C and TNM grade of the cancer, suggesting a potential value of using this gene as a prognostic marker of colorectal carcinoma. This is an interesting study.
Peer reviewer: Jian-Yuan Chai, PhD, Director of the Laboratory of GI Injury and Cancer, Research Scientist of Department of Veterans Affairs, Assistant Professor of University of California, 5901 E. 7th St, Long Beach, CA 90822, United States
S- Editor Wen LL L- Editor Rutherford A E- Editor Li JY
| 1. | Sung JJ, Lau JY, Goh KL, Leung WK. Increasing incidence of colorectal cancer in Asia: implications for screening. Lancet Oncol. 2005;6:871-876. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 2. | Day JA, Mason RR, Chesrown SE. Effect of massage on serum level of beta-endorphin and beta-lipotropin in healthy adults. Phys Ther. 1987;67:926-930. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 34] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 3. | Shastry BS. SNP alleles in human disease and evolution. J Hum Genet. 2002;47:561-566. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 247] [Cited by in F6Publishing: 233] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 4. | Park JH, Gail MH, Greene MH, Chatterjee N. Potential usefulness of single nucleotide polymorphisms to identify persons at high cancer risk: an evaluation of seven common cancers. J Clin Oncol. 2012;30:2157-2162. [PubMed] [Cited in This Article: ] |
| 5. | Gaj P, Maryan N, Hennig EE, Ledwon JK, Paziewska A, Majewska A, Karczmarski J, Nesteruk M, Wolski J, Antoniewicz AA. Pooled sample-based GWAS: a cost-effective alternative for identifying colorectal and prostate cancer risk variants in the Polish population. PLoS One. 2012;7:e35307. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 31] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 6. | Dai J, Wan S, Zhou F, Myers RE, Guo X, Li B, Fu X, Palazzo JP, Dou K, Yang H. Genetic polymorphism in a VEGF-independent angiogenesis gene ANGPT1 and overall survival of colorectal cancer patients after surgical resection. PLoS One. 2012;7:e34758. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 13] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 7. | Gessner R, Tauber R. Intestinal cell adhesion molecules. Liver-intestine cadherin. Ann N Y Acad Sci. 2000;915:136-143. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 81] [Cited by in F6Publishing: 80] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 8. | Kreft B, Berndorff D, Böttinger A, Finnemann S, Wedlich D, Hortsch M, Tauber R, Gessner R. LI-cadherin-mediated cell-cell adhesion does not require cytoplasmic interactions. J Cell Biol. 1997;136:1109-1121. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 71] [Cited by in F6Publishing: 80] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 9. | Nagafuchi A, Takeichi M. Cell binding function of E-cadherin is regulated by the cytoplasmic domain. EMBO J. 1988;7:3679-3684. [PubMed] [Cited in This Article: ] |
| 10. | Hinoi T, Lucas PC, Kuick R, Hanash S, Cho KR, Fearon ER. CDX2 regulates liver intestine-cadherin expression in normal and malignant colon epithelium and intestinal metaplasia. Gastroenterology. 2002;123:1565-1577. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 163] [Cited by in F6Publishing: 168] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 11. | Grötzinger C, Kneifel J, Patschan D, Schnoy N, Anagnostopoulos I, Faiss S, Tauber R, Wiedenmann B, Gessner R. LI-cadherin: a marker of gastric metaplasia and neoplasia. Gut. 2001;49:73-81. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 12. | Mokrowiecka A, Zonnur S, Veits L, Musial J, Kordek R, Lochowski M, Kozak J, Malecka-Panas E, Vieth M, Hartmann A. Liver-Intestine-Cadherin Is a Sensitive Marker of Intestinal Differentiation During Barrett's Carcinogenesis. Dig Dis Sci. 2012;Oct 4; Epub ahead of print. [PubMed] [Cited in This Article: ] |
| 13. | Papadopoulou E, Simopoulos K, Tripsianis G, Tentes I, Anagnostopoulos K, Sivridis E, Galazios G, Kortsaris A. Allelic imbalance of HER-2 codon 655 polymorphism among different religious/ethnic populations of northern Greece and its association with the development and the malignant phenotype of breast cancer. Neoplasma. 2007;54:365-373. [PubMed] [Cited in This Article: ] |
| 14. | Zheng X, Wang L, Zhu Y, Guan Q, Li H, Xiong Z, Deng L, Lu J, Miao X, Cheng L. The SNP rs961253 in 20p12.3 is associated with colorectal cancer risk: a case-control study and a meta-analysis of the published literature. PLoS One. 2012;7:e34625. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 13] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 15. | Perrone CE, Mattocks DA, Plummer JD, Chittur SV, Mohney R, Vignola K, Orentreich DS, Orentreich N. Genomic and metabolic responses to methionine-restricted and methionine-restricted, cysteine-supplemented diets in Fischer 344 rat inguinal adipose tissue, liver and quadriceps muscle. J Nutrigenet Nutrigenomics. 2012;5:132-157. [PubMed] [Cited in This Article: ] |
| 16. | Suh YS, Lee HJ, Jung EJ, Kim MA, Nam KT, Goldenring JR, Yang HK, Kim WH. The combined expression of metaplasia biomarkers predicts the prognosis of gastric cancer. Ann Surg Oncol. 2012;19:1240-1249. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 29] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 17. | Ko S, Chu KM, Luk JM, Wong BW, Yuen ST, Leung SY, Wong J. Overexpression of LI-cadherin in gastric cancer is associated with lymph node metastasis. Biochem Biophys Res Commun. 2004;319:562-568. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 31] [Cited by in F6Publishing: 34] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 18. | Yasui W, Oue N, Sentani K, Sakamoto N, Motoshita J. Transcriptome dissection of gastric cancer: identification of novel diagnostic and therapeutic targets from pathology specimens. Pathol Int. 2009;59:121-136. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 34] [Cited by in F6Publishing: 38] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 19. | Liu QS, Zhang J, Liu M, Dong WG. Lentiviral-mediated miRNA against liver-intestine cadherin suppresses tumor growth and invasiveness of human gastric cancer. Cancer Sci. 2010;101:1807-1812. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25] [Cited by in F6Publishing: 29] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 20. | Ito R, Oue N, Yoshida K, Kunimitsu K, Nakayama H, Nakachi K, Yasui W. Clinicopathological significant and prognostic influence of cadherin-17 expression in gastric cancer. Virchows Arch. 2005;447:717-722. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 38] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 21. | Wang J, Yu JC, Kang WM, Wang WZ, Liu YQ, Gu P. The predictive effect of cadherin-17 on lymph node micrometastasis in pN0 gastric cancer. Ann Surg Oncol. 2012;19:1529-1534. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 26] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 22. | Resende C, Thiel A, Machado JC, Ristimäki A. Gastric cancer: basic aspects. Helicobacter. 2011;16 Suppl 1:38-44. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 50] [Cited by in F6Publishing: 55] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 23. | Panarelli NC, Yantiss RK, Yeh MM, Liu Y, Chen YT. Tissue-specific cadherin CDH17 is a useful marker of gastrointestinal adenocarcinomas with higher sensitivity than CDX2. Am J Clin Pathol. 2012;138:211-222. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 62] [Cited by in F6Publishing: 60] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 24. | Hippo Y, Taniguchi H, Tsutsumi S, Machida N, Chong JM, Fukayama M, Kodama T, Aburatani H. Global gene expression analysis of gastric cancer by oligonucleotide microarrays. Cancer Res. 2002;62:233-240. [PubMed] [Cited in This Article: ] |
| 25. | Takamura M, Sakamoto M, Ino Y, Shimamura T, Ichida T, Asakura H, Hirohashi S. Expression of liver-intestine cadherin and its possible interaction with galectin-3 in ductal adenocarcinoma of the pancreas. Cancer Sci. 2003;94:425-430. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 2] [Reference Citation Analysis (0)] |
| 26. | Lee NP, Poon RT, Shek FH, Ng IO, Luk JM. Role of cadherin-17 in oncogenesis and potential therapeutic implications in hepatocellular carcinoma. Biochim Biophys Acta. 2010;1806:138-145. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 26] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 27. | Takamura M, Ichida T, Matsuda Y, Kobayashi M, Yamagiwa S, Genda T, Shioji K, Hashimoto S, Nomoto M, Hatakeyama K. Reduced expression of liver-intestine cadherin is associated with progression and lymph node metastasis of human colorectal carcinoma. Cancer Lett. 2004;212:253-259. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 42] [Cited by in F6Publishing: 46] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 28. | Huang LP, Yu YH, Sheng C, Wang SH. Up-regulation of cadherin 17 and down-regulation of homeodomain protein CDX2 correlate with tumor progression and unfavorable prognosis in epithelial ovarian cancer. Int J Gynecol Cancer. 2012;22:1170-1176. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 10] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 29. | Gröne J, Lenze D, Jurinovic V, Hummel M, Seidel H, Leder G, Beckmann G, Sommer A, Grützmann R, Pilarsky C. Molecular profiles and clinical outcome of stage UICC II colon cancer patients. Int J Colorectal Dis. 2011;26:847-858. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 31] [Cited by in F6Publishing: 34] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 30. | Su MC, Yuan RH, Lin CY, Jeng YM. Cadherin-17 is a useful diagnostic marker for adenocarcinomas of the digestive system. Mod Pathol. 2008;21:1379-1386. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 31. | Kwak JM, Min BW, Lee JH, Choi JS, Lee SI, Park SS, Kim J, Um JW, Kim SH, Moon HY. The prognostic significance of E-cadherin and liver intestine-cadherin expression in colorectal cancer. Dis Colon Rectum. 2007;50:1873-1880. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 41] [Cited by in F6Publishing: 46] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 32. | Wang XQ, Luk JM, Leung PP, Wong BW, Stanbridge EJ, Fan ST. Alternative mRNA splicing of liver intestine-cadherin in hepatocellular carcinoma. Clin Cancer Res. 2005;11:483-489. [PubMed] [Cited in This Article: ] |
| 33. | Wang XQ, Luk JM, Garcia-Barcelo M, Miao X, Leung PP, Ho DW, Cheung ST, Lam BY, Cheung CK, Wong AS. Liver intestine-cadherin (CDH17) haplotype is associated with increased risk of hepatocellular carcinoma. Clin Cancer Res. 2006;12:5248-5252. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 28] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 34. | Zhang J, Liu QS, Dong WG. Blockade of proliferation and migration of gastric cancer via targeting CDH17 with an artificial microRNA. Med Oncol. 2011;28:494-501. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 15] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 35. | Yu QF, Dong WG, Ren JL. Knockdown of Li-cadherin increases metastatic behaviors of LoVo cells. J Cancer Res Clin Oncol. 2010;136:1641-1649. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 7] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 36. | Park JH, Seol JA, Choi HJ, Roh YH, Choi PJ, Lee KE, Roh MS. Comparison of cadherin-17 expression between primary colorectal adenocarcinomas and their corresponding metastases: the possibility of a diagnostic marker for detecting the primary site of metastatic tumour. Histopathology. 2011;58:315-318. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 19] [Article Influence: 1.5] [Reference Citation Analysis (0)] |