Copyright
©2011 Baishideng Publishing Group Co.
World J Gastroenterol. Aug 7, 2011; 17(29): 3407-3419
Published online Aug 7, 2011. doi: 10.3748/wjg.v17.i29.3407
Published online Aug 7, 2011. doi: 10.3748/wjg.v17.i29.3407
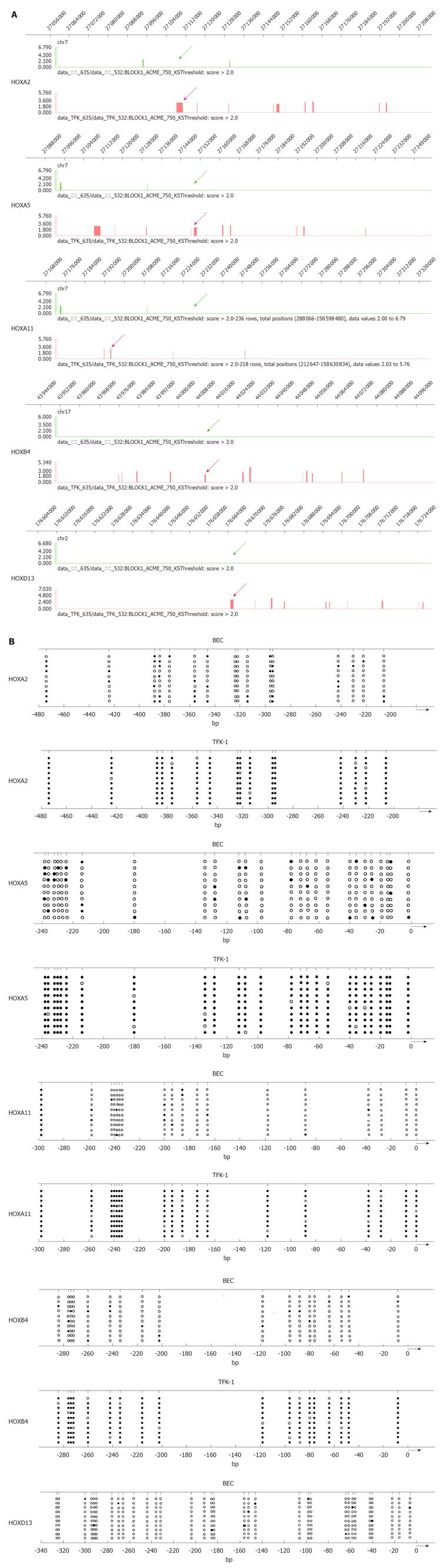
Figure 2 DNA methylation analyses of CpG islands of hypermethylated HOX genes.
A: Graphical representation of Methylated DNA Immunoprecipitation microarray (Signalmap software, NimbleGen). The panels show the DNA methylation profile at homeobox cluster genes in normal epithelial cell of bile duct cell line bile duct epithelial cells (BEC) and cholangiocarcinoma cell line TFK-1. The methylated CpG islands are indicated by bar and arrow in TFK-1 (red) and BEC (green). Chromosomal location is indicated at the top of the diagram; B: Bisulfate-sequencing of HOXA2, HOXA5, HOXA11, HOXB4 and HOXD13. Each vertical line represents a single CpG site on the top. The regions were analyzed by bisulfate-sequencing. The transcription start site and location of exon 1 are shown by thick bars on the bottom. Each row represents an individual cloned allele. Circles represent CpG sites and their spacing accurately reflects the CpG density of the region. Black circles, methylated CpG site; white circles, unmethylated CpG site. Dense methylation at the promoters was found in TFK-1. In contrast, promoter hypomethylation found in BEC among all the genes.
- Citation: Shu Y, Wang B, Wang J, Wang JM, Zou SQ. Identification of methylation profile of HOX genes in extrahepatic cholangiocarcinoma. World J Gastroenterol 2011; 17(29): 3407-3419
- URL: https://www.wjgnet.com/1007-9327/full/v17/i29/3407.htm
- DOI: https://dx.doi.org/10.3748/wjg.v17.i29.3407