Published online May 7, 2011. doi: 10.3748/wjg.v17.i17.2248
Revised: December 9, 2010
Accepted: December 16, 2010
Published online: May 7, 2011
AIM: To evaluate the association of human leukocyte antigen (HLA)-DQB1 alleles with hepatocellular carcinoma (HCC) through meta-analysis of published data.
METHODS: Case-control studies on HLA-DQB1 allele association with HCC published up to January 2010 were included in the analyses. The odds ratios (ORs) of HLA-DQB1 allele distributions in HCC patients were analyzed and compared with healthy controls. The meta-analysis software REVMAN 5.0 was applied for investigating heterogeneity among individual studies and for summarizing all the studies. A meta-analysis was performed using fixed-effect or random-effect methods, depending on the absence or presence of significant heterogeneity. Seven case-control studies containing 398 cases and 594 controls were included in the final analysis.
RESULTS: Among the five family alleles, two (DQB1*02 and DQB1*03) were found to be significantly associated with the risk of HCC. The combined OR for the association of DQB1*02 and DQB1*03 allele with the risk for HCC was 1.78 (95% CI: 1.05-3.03, P = 0.03) and 0.65 (95% CI: 0.48-0.89, P = 0.007), respectively. Among the 13 specific alleles, two (DQB1*0502 and DQB1*0602) were significantly associated with risk of HCC. The combined OR for the association of DQB1*0502 and DQB1*0602 allele with the risk for HCC was 1.82 (95% CI: 1.14-2.92, P = 0.01) and 0.58 (95% CI: 0.36-0.95, P = 0.03), respectively. No significant association was established for other HLA-DQB1 family alleles and specific alleles.
CONCLUSION: Our results support the hypothesis that specific HLA-DQB1 allele families and alleles might influence the susceptibility or resistance to HCC, although it needs further investigations.
- Citation: Xin YN, Lin ZH, Jiang XJ, Zhan SH, Dong QJ, Wang Q, Xuan SY. Specific HLA-DQB1 alleles associated with risk for development of hepatocellular carcinoma: A meta-analysis. World J Gastroenterol 2011; 17(17): 2248-2254
- URL: https://www.wjgnet.com/1007-9327/full/v17/i17/2248.htm
- DOI: https://dx.doi.org/10.3748/wjg.v17.i17.2248
Hepatocellular carcinoma (HCC) is the third most common cause of cancer-related deaths worldwide and about 600 000 patients died from the disease annually[1]. Nearly 78% of the 600 000 cases are from Asian countries[2]. China alone accounts for more than 50% of the world’s cases[3]. The development of HCC is linked to the interaction of the genetic, immunologic, environmental, dietary, and lifestyle factors. Its incidence and distribution vary widely among ethnic groups, sex, and geographic regions. HBV and HCV infection, liver cirrhosis, male gender, and old age are important risk factors of HCC. The clustering of HCC within families raises the possibility that genetic factors are also involved in the susceptibility to HCC.
The major histocompatibility complex (MHC) plays a key role in anti-virus and tumor defense. Human leukocyte antigens (HLA) function in the regulation of immune response to foreign antigens and discrimination of self from non-self antigens. They are encoded by a series of closely linked genetic loci found on chromosome 6[4,5]. HLA polymorphism is implicated in conferring genetic susceptibility to a large number of immune-mediated diseases, including some cancers. Given the pivotal role of HLA molecules in the immune system, several studies have investigated the association between specific HLA alleles and HCC. However, the association between HLA-DQB1 alleles and HCC in different ethnic populations is controversial. As many conflicting reports have been published to date, we conducted a systematic review of all the relevant studies published in the literature to evaluate the association between HLA-DQB1 alleles and HCC. Our principal objectives were to clarify the specific HLA-DQB1 allele families or alleles that are associated with the risk of HCC development.
Electronic databases (PubMed, EMBASE, Cochrane Library and China National Knowledge Infrastructure) were used to search for all genetic association studies evaluating the HLA-DQB1 polymorphism and HCC in humans in all languages up to January 2010. The search strategy was based on combinations of the key words, “HLA” or “human leukocyte antigen” and “hepatocellular carcinoma” or “HCC”, and was not restricted by period. We also did a full manual search from the bibliographies of selected papers. In addition, we contacted the authors of studies containing relevant information, who did not report the results necessary for this analysis. Unpublished data were also accepted if an abstract was available and further information was obtained from the authors.
In the meta-analysis, the following inclusion criteria were set and reviewed by two independent investigators: (1) an independent case-control study; (2) studies with similar purpose and statistical methods; (3) studies providing enough information to calculate the odds ratio (OR); (4) HLA-DQB1 alleles were molecularly typed (high or low resolution level); and (5) the diagnosis of HCC was based on at least one of the following criteria: typical histological characteristics or serum α-fetoprotein (AFP) levels higher than 400 ng/mL together with radiological findings (ultrasound and CT) consistent with HCC.
The following exclusion criteria were set: (1) incomplete raw data; (2) repetitive reports (if more than one version of the same study was retrieved, only the most recent was used); and (3) materials and methods were not well-described and reliable.
Although assessment of study quality is considered important for systematic reviews and meta-analyses, scoring methods have been considered problematic[6] and may not accurately assess the quality measures of interest[7]. Therefore, we used reliability of patient selection, molecular typing method, and statistical analysis method as quality variables.
The frequency of HLA-DQB1 alleles varies according to ethnic and racial background, with some alleles being extremely rare. Therefore, articles were not required to identify all alleles for inclusion.
The studies were independently evaluated by two researchers (Xin YN and Lin ZH). Discrepancies in the evaluations of studies were resolved by discussion between the reviewers.
The following data were collected from each study: authors, publication year, journal, publication type and language, HLA genotyping method, allele genotype, number of cases and controls, definitions used for HCC, HCC sample description, control sample description (if there was more than one control group, we choose the healthy group as the control group in order to minimize the confounder). The main features of the trials included in the meta-analysis are shown in Table 1.
| Study | Country/region | No. of HCC (M/F), age (yr) | No. of controls (M/F), age (yr) | No. of DQB1 alleles studied | Type of control | HLA genotyping method |
| Li et al[10], 1995 | Hong Kong | 49 (-/-), ND | 78 (-/-), ND | 11 | Healthy | PCR-SSP |
| Donaldson et al[11], 2001 | Hong Kong | 84 (79/5), 55 | 124 (-/-), ND | 10 | Healthy | PCR-SSOP |
| De Re et al[12], 2004 | Italy | 29 (-/-), ND | 144 (-/-), ND | 5 | Healthy | PCR-SSP |
| López-Vázquez et al[13], 2004 | Spain | 46 (27/19), 62 ± 8 | 48 (19/29), 56 ± 12 | 10 | HCV carriers | PCR-SSOP |
| Liu et al[14], 2007 | China | 78 (63/15), 58.8 ± 11.5 | 100 (-/-), ND | 19 | Healthy | PCR-SSP |
| El-Chennawi et al[15], 2008 | Egypt | 50 (45/5), 51.16 ± 6.16 | 50 (44/6), 48.88 ± 9.22 | 5 | Healthy | PCR-SSP |
| Pan[16], 2009 | China | 62 (52/10), 53.58 | 50 (29/21), 30.12 | 8 | Healthy | PCR-SSP |
Homogeneity was calculated by Cochran’s Q test (α = 0.05). If the results of the Q test had no significant heterogeneity, the Mantel-Haenszel fixed-effect model (Peto method) was used for the combined data. If the results of the Q test had significant heterogeneity, the Dersimonian-Laird random-effects model (DL method) was used for the combination of data[8]. A pooled OR was presented as a standard plot with 95% CIs. In the absence of heterogeneity, the two methods provided identical results. As a measure of association between HCC and HLA-DQB1 alleles, we combined ORs with 95% CIs stratified by gene subtype of patients and controls in a study. Funnel plots and the Egger’s regression asymmetry test were used to evaluate publication bias[9]. We performed a sensitivity analysis to assess the stability of the results by sequential omission of individual studies. All P values presented are two-tailed. The analyses were performed using Revman 5.0 provided by the Cochrane Collaboration Internet.
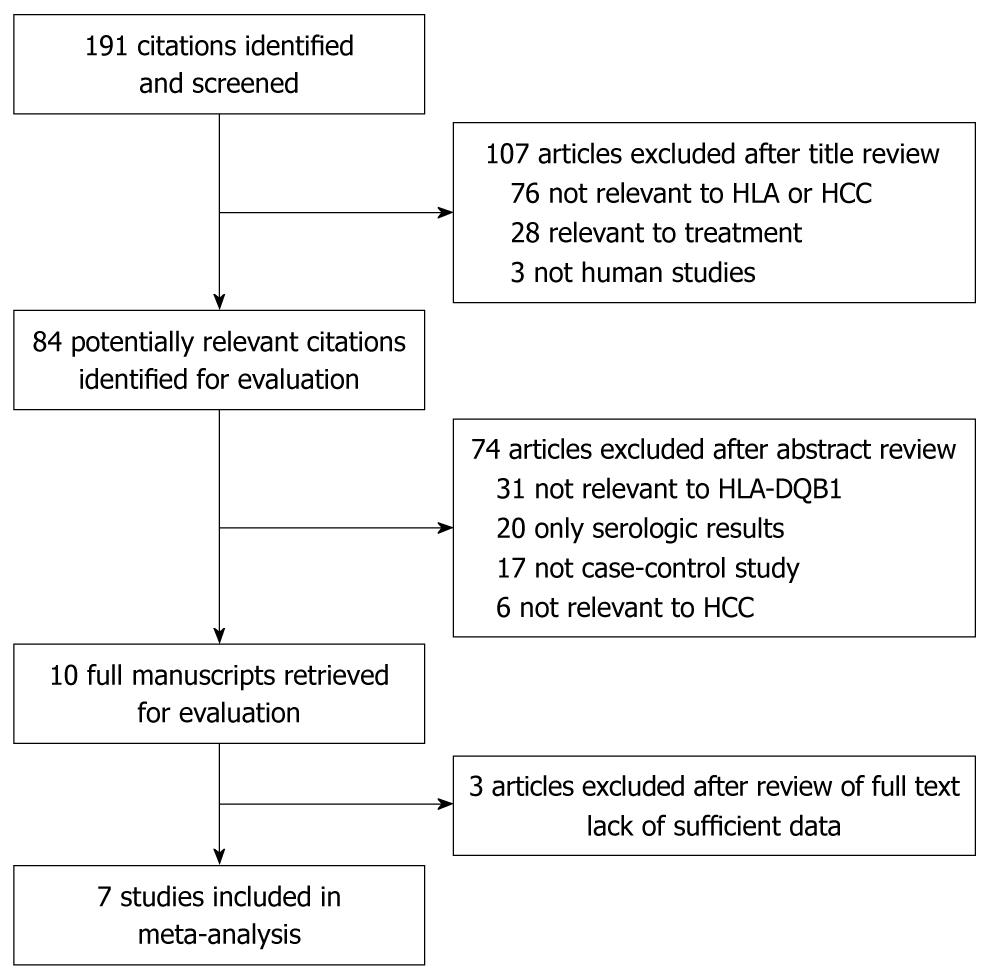
Figure 1 shows the flow diagram of publications identified by the literature search. The search strategy allowed us to identify 60 studies for potential inclusion in the meta-analysis. Only 7 case-control studies relating to HLA-DQB1 alleles polymorphism and susceptibility to HCC qualified on the basis of our selection criteria[10-16]. A total of 991 subjects were studied (398 patients and 593 controls) (Table 1). Two of these studies were conducted in mainland China, two in Hong Kong, and one each in Italy, Egypt, and Spain. The diagnosis of HCC was based on at least one of the following criteria: typical histological characteristics or serum AFP levels higher than 400 ng/mL together with radiological findings (ultrasound and/or CT) consistent with HCC. The control group of six studies consisted of healthy individuals, while only one study used HCV carriers as the control group[13]. Records of mean or median age and sex were incomplete in 4/7 reports. HIV status was determined in two reports[12,15]. HLA-DQB1 alleles were considered only when they were classified in at least two independent studies.
HLA-DQB1 alleles were molecularly typed (high or low resolution level). Six studies used high resolution molecular typing for HLA, while the Italian study used both high and low resolution molecular typing for HLA. Low resolution molecular typing methods for HLA could not identify the specific alleles. Accurate methods for HLA class II typing should involve the combination of polymerase chain reaction (PCR)-sequence-specific oligonucleotides probes, PCR-sequence specific primer and PCR-single strand conformation polymorphism[17].
A total of five HLA-DQB1 allele families and 13 specific alleles were extracted from the studies to investigate their association with HCC. Six specific alleles were excluded because each was identified in only one study.
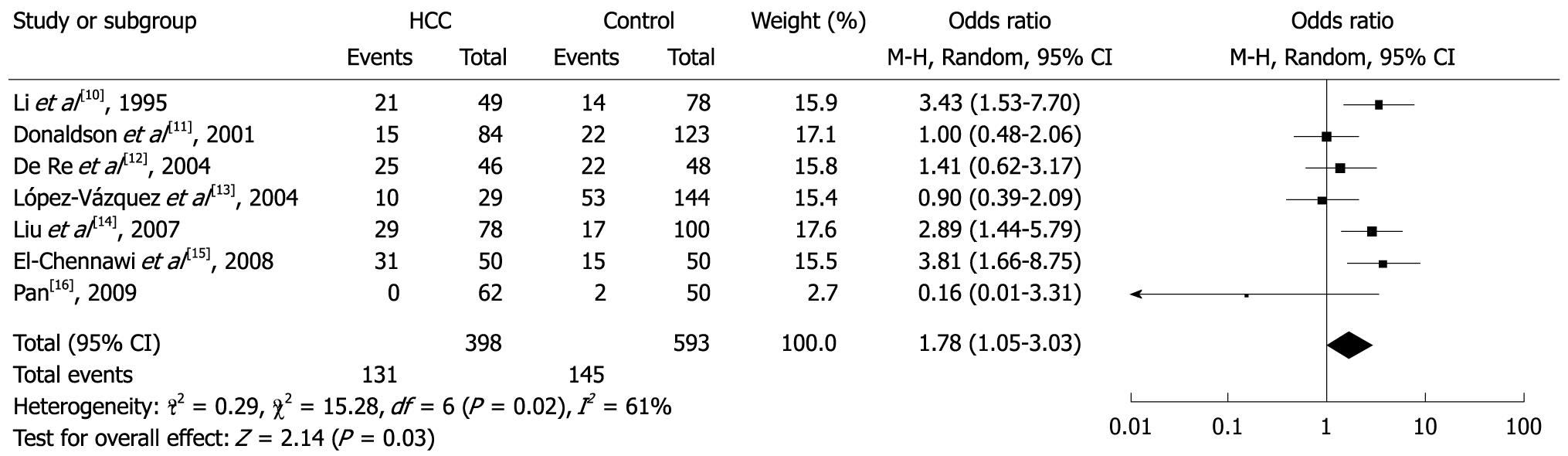
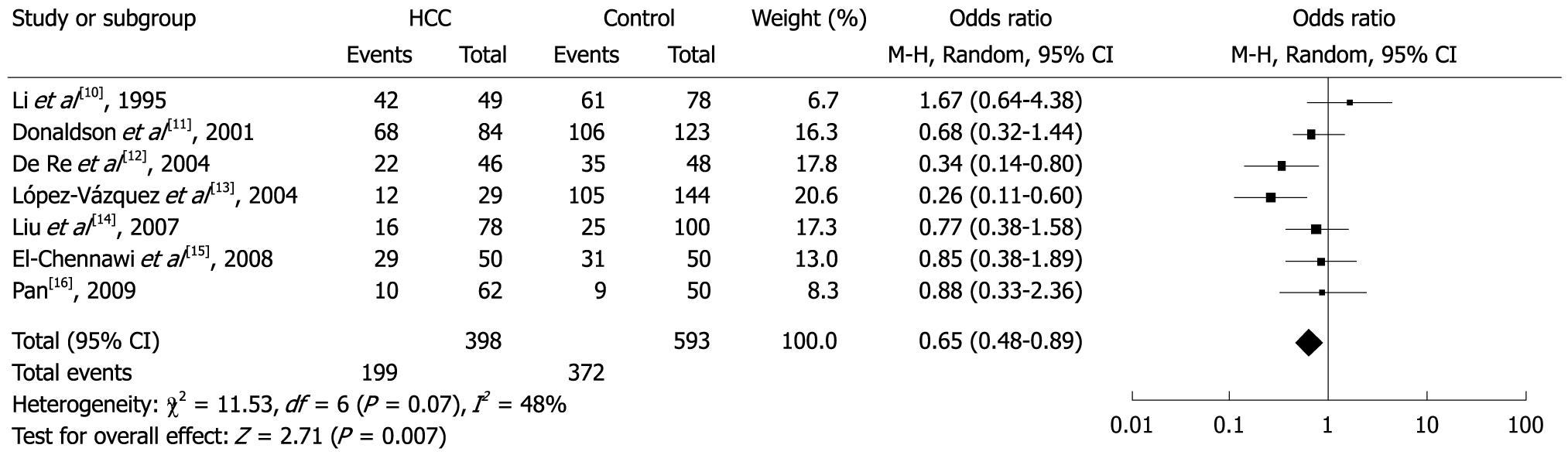
Among the allele families, two (DQB1*02 and DQB1*03) were significantly associated with risk of HCC. In the meta-analysis, the overall frequency of DQB1*02 was 32.9% (131/398) in HCC, and 24.5% (145/593) in controls. The heterogeneity test indicated that the variation of trial-specific ORs was statistically significant (χ2 = 15.28, P = 0.02 and < 0.05); the random-effect method was used to combine the results. The combined OR was 1.78 (95% CI: 1.05-3.03) and was statistically significant (P = 0.03 and < 0.05). In sensitivity analysis, the exclusion of individual studies did not change this significant result, except for the studies by Li et al[10], López-Vázquez et al[13], Liu et al[14] and El-Chennawi et al[15], which produced a non-significant association. Overall, the frequency of DQB1*03 was 50.0% (199/398) in HCC and 62.7% (372/593) in controls. The heterogeneity test indicated that the variation of trial-specific ORs was not statistically significant (χ2 = 11.53, P = 0.07 and > 0.05). The fixed-effect method was used to combine the results. The combined OR was 0.65 (95% CI: 0.48-0.89) and was statistically significant (P = 0.007 and < 0.05). In sensitivity analysis, the exclusion of individual studies did not change this significant result, except for the study by De Re et al[12], which produced a non-significant association. Statistics calculated for each study about DQB1*03 are shown in the forest plot (Figures 2 and 3). Meta-analysis for another 3 HLA-DQB1 allele families (DQB1*04, DQB1*05 and DQB1*06) was carried out, but did not show any statistical effect.
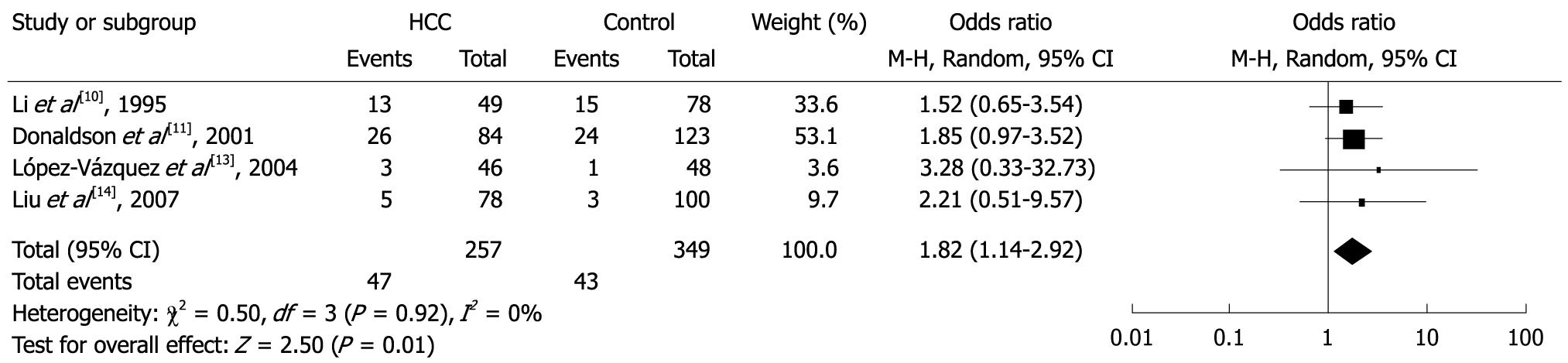
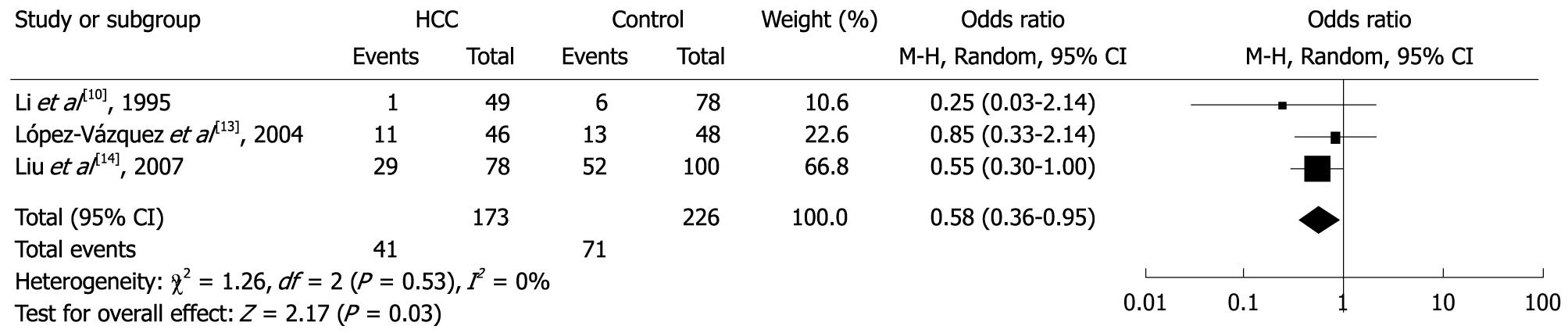
Among the specific alleles, two (DQB1*0502 and DQB1*0602) were significantly associated with risk of HCC. In the meta-analysis, the overall frequency of HLA-DQB1*0502 allele was 18.3% (47/257) in HCC, and 12.3% (43/349) in the controls. The heterogeneity test indicated that the variation of trial-specific ORs was not statistically significant (χ2 = 0.50, P = 0.92 and > 0.05); the fixed-effect method was used to combine the results. The combined OR was 1.82 (95% CI: 1.14-2.92), and was statistically significant (P = 0.01 and < 0.05). In sensitivity analysis, the exclusion of individual studies did not change this significant result, except for the study by Donaldson et al[11], which produced a non-significant association. Overall, the frequency of HLA- DQB1*0602 allele was 23.7% (41/173) in HCC and 31.4% (71/226) in controls. The heterogeneity test indicated that the variation of trial-specific ORs was not statistically significant (χ2 = 1.26, P = 0.53 and > 0.05); the fixed-effect method was used to combine the results. The combined OR was 0.58 (95% CI: 0.36-0.95) and was statistically significant (P = 0.03 and < 0.05). In sensitivity analysis, the exclusion of individual studies did not change this significant result, except for the study by Liu et al[14], which produced a non-significant association. Statistics calculated for each study of DQB1*0502 and DQB1*0602 are shown in the forest plot (Figures 4 and 5). Meta-analysis for another 11 HLA-DQB1 specific alleles (DQB1*0201, DQB1*0301, DQB1*0302, DQB1*0303, DQB1*0401, DQB1*0402, DQB1*0501, DQB1*0503, DQB1*0601, DQB1*0603 and DQB1*0604) was carried out, but did not show any statistical relationships.
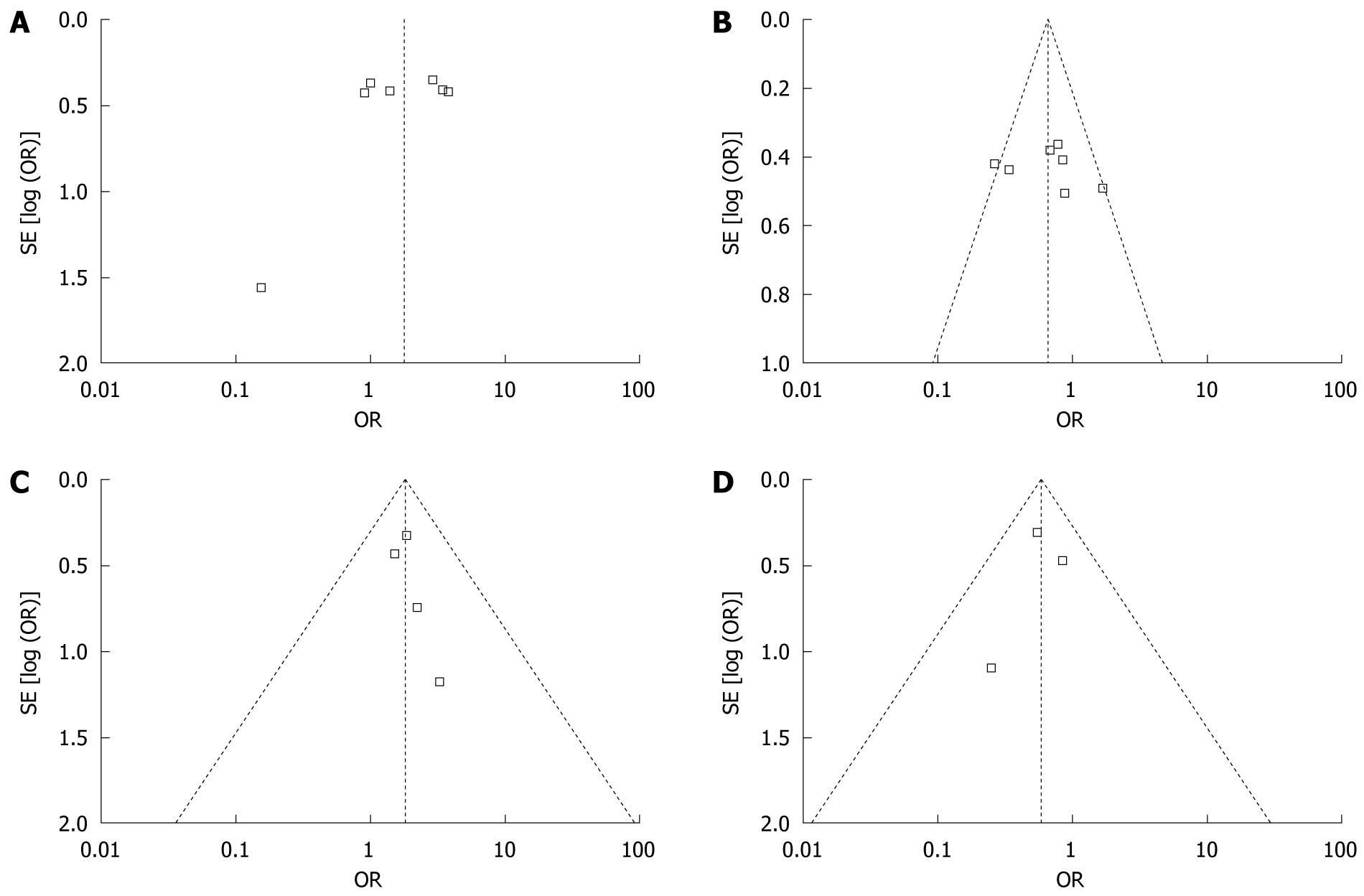
Our meta-analysis of seven studies revealed that DQB1*02 and DQB1*0502 were risk factors for HCC, while DQB1*03 and DQB1*0602 were protective factors. These results suggest that patients with DQB1*02 and DQB1*0502 alleles were at a higher risk of developing HCC than those with DQB1*03 and DQB1*0602 alleles. These analyses were based on the data from seven studies irrespective of the ethnicity of the study populations. The shape of the funnel plot to detect publication bias of each study for DQB1*02, DQB1*03, DQB1*0502 and DQB1*0602 seemed to be asymmetrical (Figure 6), suggesting that publication bias might affect the findings of our meta-analysis.
To our knowledge, this is the first published meta-analysis to comprehensively investigate the association between HLA-DQB1 alleles and HCC. Numerous studies have reported associations of HLA with malignant tumors, such as thyroid cancer, renal cell cancer, Burkett’s lymphoma, Kaposi’s sarcoma, Hodgkin’s disease, nasopharyngeal carcinoma, cervical carcinoma, esophageal carcinoma, gastric cancer, and HCC. However, most of these studies focused on the association between HLA antigen and cancer based on HLA serotyping which has limited resolution, and may be inaccurate for the assignment of many HLA antigens. With the development of DNA-based methods for HLA genotyping, HLA genotyping has become more accurate in the determination of the HLA-DQB1 alleles, and more precise than serotyping in the determination of the peptide-binding domain of MHC class II molecules. Hence, HLA genotyping is being used more frequently in the field of immunogenetics.
Recent studies on the association between HLA-DQB1 allele polymorphisms and HCC have been inconclusive. Pan[16] reported that some HLA-DQB1 alleles are significantly associated with HCC (DRB1*04 and DQB1*02), while others (DQB1*06) are not. Therefore, it is possible that the DRB1*04 and DQB1*02 alleles might be risk factors for the occurrence of HCC (OR = 4.373 and 3.807, respectively), and DQB1*06 may be a protective allele (OR = 0.259). Lopez-Vazquez et al[13] confirmed that HLA-DQB1*0301 carriers rarely develop end-stage liver disease (ESLD). This allele may, therefore, be considered to be a good prognostic factor. De Re et al[12] have shown that the DQB1*0301 allele played a major protective role in patients with HCV-associated HCC. Donaldson et al[11] observed that the alleles DRB1*1501, DQA1*0302 and to a lesser extent DQB1*0302 appeared to confer resistance to HCC.
The current meta-analysis indicates that specific HLA-DQB1 alleles are associated with HCC. Among the 5 family alleles, DQB1*02 and DQB1*03 were significantly associated with risk of HCC. The combined OR was 1.78 (95% CI: 1.05-3.03) and 0.65 (95% CI: 0.48-0.89), respectively. Among the 13 specific alleles, DQB1*0502 and DQB1*0602 were significantly associated with risk of HCC. The combined OR was 1.82 (95% CI: 1.14-2.92) and 0.58 (95% CI: 0.36-0.95), respectively. These results suggest that DQB1*02 and DQB1*0502 alleles may have a higher risk for HCC, while DQB1*03 and DQB1*0602 alleles may have a protective effect for HCC.
The DQB1*02 allele family is negatively associated with the incidence of breast cancer in the Tunisian population[18], while the DQB1*03 allele family is associated with susceptibility to cervical carcinoma in Europe (OR = 3.03)[19], cutaneous T-cell lymphoma in USA (OR = 2.7)[20], and HCV-related non-Hodgkin’s lymphoma in Italy (OR = 0.23)[12]. The DQB1*0502 allele is also associated with the susceptibility to Sezary syndrome of cutaneous T-cell lymphoma in USA (OR = 7.75)[20], while the DQB1*0602 allele is associated with an increased gastric cancer risk in Taiwan (OR = 2.79)[21] and increased distal gastric cancer risk in southern European population (OR = 4.82)[22]. DQB1*0602 also appears to be a candidate protective allele for testicular germ cell carcinoma in the Japanese population (RR = 0.26, P = 0.02)[23]. The mechanisms for these effects are unknown, but larger-scale studies may confirm this observation.
Human tumor cells express diverse types of antigens, depending on the etiology and pathogenesis of the disease[24]. Because tumor development is preceded by chronic inflammation, immune responses, whether towards the infectious agent itself or against tumor antigens, may be critical for development of tumor. Different types of HLA molecules have different capability of binding and presenting tumor antigens. The association of specific HLA alleles with susceptibility or resistance to malignant tumors is probably attributable to a direct involvement of the HLA molecules as an antigen presenter or possibly due to a neighboring linked gene[14].
Several other points should be considered when interpreting the results of our study. Most studies did not control for the matching variables in the analysis, and the risk for HCC was not controlled for possible confounders such as HBV or HCV. These observational studies are more prone to bias than randomized clinical trial (RCT) studies. The shape of the funnel plot seemed to be asymmetrical, suggesting that publication bias might affect the findings of our meta-analysis.
In conclusion, this meta-analysis reveals that DQB1*02 and DQB1*0502 are risk factors for HCC, while DQB1*03 and DQB1*0602 are protective factors. These findings are consistent with the previous studies about the association of HLA alleles and other cancers. In the present study, no attempt has been made to account for the potential confounding influence of cirrhosis in the development of HCC, although future studies of HLA should take this into consideration.
Hepatocellular carcinoma (HCC) is the third most common cause of cancer-related deaths worldwide and about 600 000 patients died from the disease annually. Human leukocyte antigen (HLA) polymorphism is implicated in conferring genetic susceptibility to a large number of immune-mediated diseases, including some cancers.
The clustering of HCC within families raises the possibility that genetic factors are involved in susceptibility to HCC. Much attention has been paid to the potential role of HLA in the pathogenesis of HCC in an attempt to uncover the underlying mechanisms. However, the relationship between HLA-DQB1 and HCC remains controversial and no meta-analysis has been conducted.
This is the first meta-analysis which systemically studied the relationship between HLA-DQB1 and HCC susceptibility, and suggested that specific HLA-DQB1 allele families and alleles might influence the susceptibility or resistance to HCC, which needs further investigations.
The study concludes that DQB1*02 and DQB1*0502 are risk factors for HCC, while DQB1*03 and DQB1*0602 are protective factors, which provides an insight into the role of HLA-DQB1 in the pathogenesis of HCC. Furthermore, these findings are meaningful to early diagnosis and provide a new strategic approach to the prevention of HCC.
Meta-analysis is a means of increasing the effective sample size under investigation through the pooling of data from individual association studies, thus enhancing the statistical power of the analysis.
This is a very interesting meta-analytic study dealing with an important topic in HCC. The described analysis has been performed with precision and accuracy.
Peer reviewer: Jian Wu, Associate Professor of Medicine, Internal Medicine/Transplant Research Program, University of California, Davis Medical Center, 4635 2nd Ave. Suite 1001, Sacramento CA 95817, United States
S- Editor Sun H L- Editor Ma JY E- Editor Zheng XM
| 1. | Schütte K, Bornschein J, Malfertheiner P. Hepatocellular carcinoma--epidemiological trends and risk factors. Dig Dis. 2009;27:80-92. [Cited in This Article: ] |
| 2. | Keeffe EB, Dieterich DT, Pawlotsky JM, Benhamou Y. Chronic hepatitis B: preventing, detecting, and managing viral resistance. Clin Gastroenterol Hepatol. 2008;6:268-274. [Cited in This Article: ] |
| 3. | El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557-2576. [Cited in This Article: ] |
| 4. | Klein J, Sato A. The HLA system. First of two parts. N Engl J Med. 2000;343:702-709. [Cited in This Article: ] |
| 5. | Klein J, Sato A. The HLA system. Second of two parts. N Engl J Med. 2000;343:782-786. [Cited in This Article: ] |
| 6. | Conn VS, Rantz MJ. Research methods: managing primary study quality in meta-analyses. Res Nurs Health. 2003;26:322-333. [Cited in This Article: ] |
| 7. | Huwiler-Müntener K, Jüni P, Junker C, Egger M. Quality of reporting of randomized trials as a measure of methodologic quality. JAMA. 2002;287:2801-2804. [Cited in This Article: ] |
| 8. | DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177-188. [Cited in This Article: ] |
| 9. | Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629-634. [Cited in This Article: ] |
| 10. | Li PK, Leung NW, Poon AS, Wong KC, Chan TH, Lai KN. Molecular genetics of major histocompatibility complex class II genes in hepatocellular carcinoma. Dig Dis Sci. 1995;40:1542-1546. [Cited in This Article: ] |
| 11. | Donaldson PT, Ho S, Williams R, Johnson PJ. HLA class II alleles in Chinese patients with hepatocellular carcinoma. Liver. 2001;21:143-148. [Cited in This Article: ] |
| 12. | De Re V, Caggiari L, Talamini R, Crovatto M, De Vita S, Mazzaro C, Cannizzaro R, Dolcetti R, Boiocchi M. Hepatitis C virus-related hepatocellular carcinoma and B-cell lymphoma patients show a different profile of major histocompatibility complex class II alleles. Hum Immunol. 2004;65:1397-404. [Cited in This Article: ] |
| 13. | López-Vázquez A, Rodrigo L, Miña-Blanco A, Martínez-Borra J, Fuentes D, Rodríguez M, Pérez R, González S, López-Larrea C. Extended human leukocyte antigen haplotype EH18.1 influences progression to hepatocellular carcinoma in patients with hepatitis C virus infection. J Infect Dis. 2004;189:957-963. [Cited in This Article: ] |
| 14. | Liu C, Cheng B. Association of polymorphisms of human leucocyte antigen-DQA1 and DQB1 alleles with chronic hepatitis B virus infection, liver cirrhosis and hepatocellular carcinoma in Chinese. Int J Immunogenet. 2007;34:373-378. [Cited in This Article: ] |
| 15. | El-Chennawi FA, Auf FA, Metwally SS, Mosaad YM, El-Wahab MA, Tawhid ZE. HLA-class II alleles in Egyptian patients with hepatocellular carcinoma. Immunol Invest. 2008;37:661-674. [Cited in This Article: ] |
| 16. | Pan HF. Association of gene polymorphism and expression of human leukocyte antigen-DRB1, DQB1 with hepatocellular carcinoma. JiLin: JiLin University 2009; . [Cited in This Article: ] |
| 17. | Thio CL, Thomas DL, Carrington M. Chronic viral hepatitis and the human genome. Hepatology. 2000;31:819-827. [Cited in This Article: ] |
| 18. | Baccar Harrath A, Yacoubi Loueslati B, Troudi W, Hmida S, Sedkaoui S, Dridi A, Jridi A, Ben Ayed F, Ben Rhomdhane K, Ben Ammar Elgaaied A. HLA class II polymorphism: protective or risk factors to breast cancer in Tunisia? Pathol Oncol Res. 2006;12:79-81. [Cited in This Article: ] |
| 19. | Odunsi K, Terry G, Ho L, Bell J, Cuzick J, Ganesan TS. Association between HLA DQB1 * 03 and cervical intra-epithelial neoplasia. Mol Med. 1995;1:161-171. [Cited in This Article: ] |
| 20. | Jackow CM, McHam JB, Friss A, Alvear J, Reveille JR, Duvic M. HLA-DR5 and DQB1*03 class II alleles are associated with cutaneous T-cell lymphoma. J Invest Dermatol. 1996;107:373-376. [Cited in This Article: ] |
| 21. | Wu MS, Hsieh RP, Huang SP, Chang YT, Lin MT, Chang MC, Shun CT, Sheu JC, Lin JT. Association of HLA-DQB1*0301 and HLA-DQB1*0602 with different subtypes of gastric cancer in Taiwan. Jpn J Cancer Res. 2002;93:404-410. [Cited in This Article: ] |
| 22. | Quintero E, Pizarro MA, Rodrigo L, Piqué JM, Lanas A, Ponce J, Miño G, Gisbert J, Jurado A, Herrero MJ. Association of Helicobacter pylori-related distal gastric cancer with the HLA class II gene DQB10602 and cagA strains in a southern European population. Helicobacter. 2005;10:12-21. [Cited in This Article: ] |
| 23. | Ozdemir E, Kakehi Y, Mishina M, Ogawa O, Okada Y, Ozdemir D, Yoshida O. High-resolution HLA-DRB1 and DQB1 genotyping in Japanese patients with testicular germ cell carcinoma. Br J Cancer. 1997;76:1348-1352. [Cited in This Article: ] |
| 24. | Jäger D, Jäger E, Knuth A. Immune responses to tumour antigens: implications for antigen specific immunotherapy of cancer. J Clin Pathol. 2001;54:669-674. [Cited in This Article: ] |