Published online Feb 14, 2010. doi: 10.3748/wjg.v16.i6.755
Revised: November 26, 2009
Accepted: December 3, 2009
Published online: February 14, 2010
AIM: To investigate the prevalence and time of Dickkopf (DKK) family methylation and its clinical significance in hepatocarcinogenesis.
METHODS: Methylation of DKK family genes was quantitatively analyzed in 115 liver tissue samples, including 50 pairs of primary hepatocellular carcinoma (HCC) and matched noncancerous cirrhotic tissue samples, as well as 15 liver cirrhosis biopsy samples.
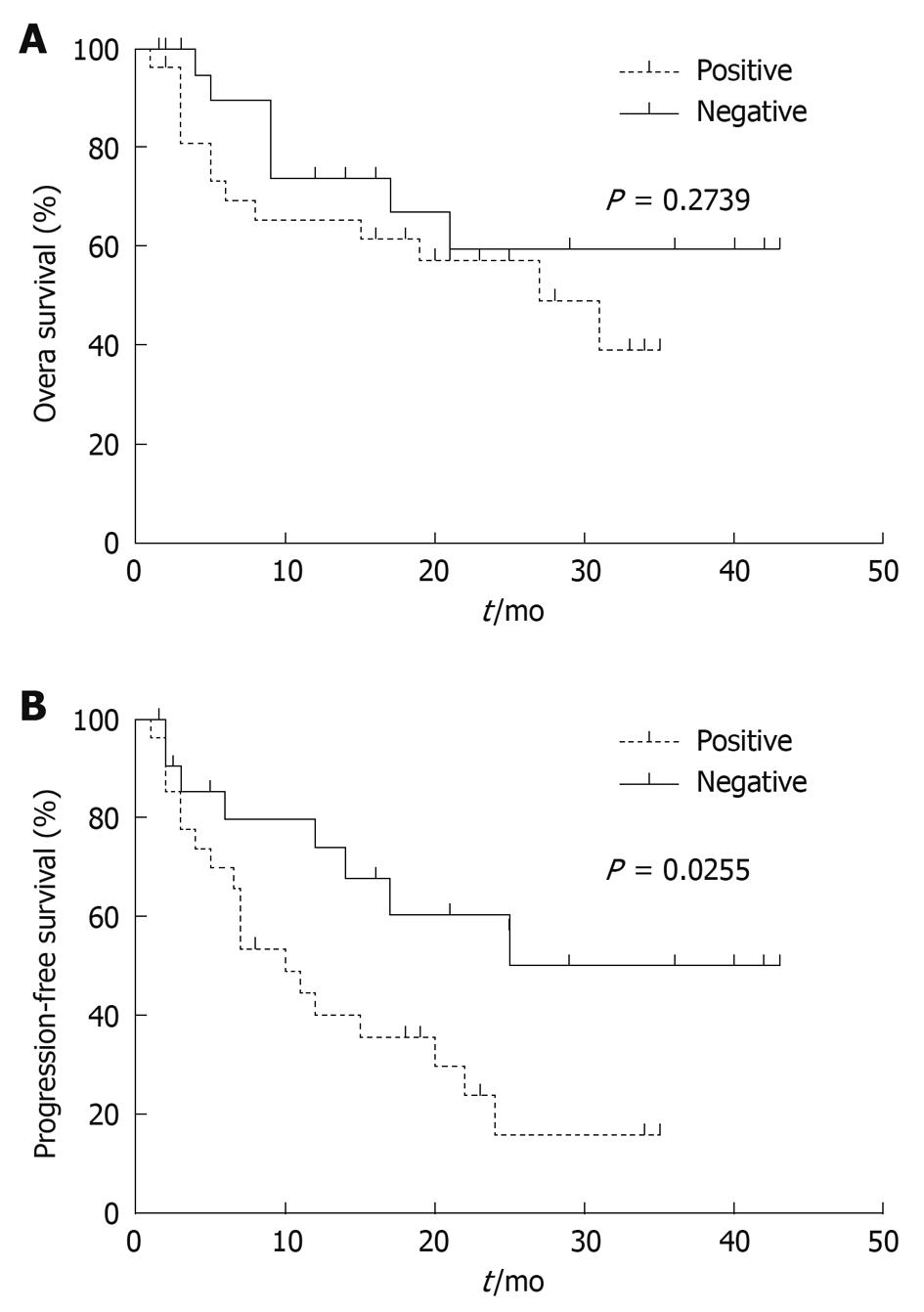
RESULTS: The methylation level of DKK3 was significantly higher in HCC tissue samples than in matched noncancerous cirrhotic tissue samples (P < 0.0001) or in liver cirrhosis biopsy samples (P = 0.0139). Receiver operator characteristic curve analysis confirmed that the percent of methylated reference (PMR) values of DKK3 could effectively discriminate HCC tissue samples from noncancerous tissue samples (AUC = 0.8146) or liver cirrhosis biopsy samples (AUC = 0.7093). Kaplan-Meier survival curves revealed that the progression-free survival time of patients with a higher DKK3 methylation level (PMR > 1%) was significantly shorter than that of those with a lower DKK3 methylation level (PMR ≤ 1%) (P = 0.0255). Multivariate Cox analysis indicated that methylated DKK3 was significantly and independently related with a shorter survival time (relative risk = 2.527, 95% CI: 1.063-6.008, P = 0.036) of HCC patients.
CONCLUSION: Methylation of DKK3 is an important event in early malignant transformation and HCC progression, and therefore might be a prognostic indicator for risk assessment of HCC.
-
Citation: Yang B, Du Z, Gao YT, Lou C, Zhang SG, Bai T, Wang YJ, Song WQ. Methylation of
Dickkopf-3 as a prognostic factor in cirrhosis-related hepatocellular carcinoma. World J Gastroenterol 2010; 16(6): 755-763 - URL: https://www.wjgnet.com/1007-9327/full/v16/i6/755.htm
- DOI: https://dx.doi.org/10.3748/wjg.v16.i6.755
Hepatocellular carcinoma (HCC) is one of the most common malignancies in the world, leading to more than 500 000 deaths every year[1]. Most HCC cases (> 80%) occur in either sub-Saharan Africa or Eastern Asia. HCC cases in China alone account for more than 40% of all cases in the world[2]. It was recently reported that the incidence of HCC is increasing worldwide[3]. Treatment of HCC is still a great challenge for clinical oncologists because most HCC patients are diagnosed at its advanced stage with distant metastasis[4].
Dysplastic cirrhotic nodules are considered precursors of HCC because of their association with HCC occurrence. Development of HCC is closely associated with cirrhosis and 90% of the tumors are found in chronic hepatitis or cirrhotic patients[5]. One reasonable explanation for this close correlation is that necrosis and regeneration of hepatocytes due to chronic liver damage provide the stepwise accumulation of genetic and epigenetic changes necessary for hepatocarcinogenesis[2]. Therefore, elucidation of these aberrant alterations involving hepatocellular transformation at the cirrhosis stage is not only crucial to understand the molecular basis of hepatocarcinogenesis but also to provide potentially useful markers for the early diagnosis, risk assessment, treatment, and chemoprevention of HCC.
Aberrant promoter hypermethylation of tumor suppressor genes is a common event during the pathogenesis of human cancers and one of the important epigenetic mechanisms in carcinogenesis. It has been shown that methylation of multiple tumor suppressor genes in HCC may contribute to the pathogenesis of this disease[5,6]. Dickkopf (DKK) family is one class of the secreted Wnt antagonists and its functional loss can contribute to activation of the Wnt pathway and result in carcinogenesis through dysregulation of cell proliferation and differentiation[7]. It has been recently shown that methylation of DKK gene family contributes to carcinogenesis and serves as a potential biomarker for the diagnosis or prognosis of several human malignancies[8]. However, few reports are available on the epigenetic silencing of DKK gene and its clinical significance in HCC[9].
In the present study, we examined the promoter hypermethylation of human DKK family genes in HCC and cirrhosis tissue samples by quantitative methylation-specific polymerase chain reaction (Q-MSP), and evaluated whether quantitative methylation of DKK genes can serve as a potentially diagnostic or prognostic biomarker for HCC.
A total of 115 liver tissue samples, including 50 pairs of primary HCC and matched noncancerous cirrhotic liver (NCL) tissue samples, as well as 15 liver cirrhotic (LC) biopsy samples, were analyzed in this study. Tumor tissue samples were collected from patients who underwent surgery in Third Central Hospital of Tianjin between December 2003 and August 2006 and stored at -80°C for further processing. Clinicopathological data were collected from patient records and pathology reports. Written informed consent was obtained from each patient and the study protocol was approved by the Clinical Research Ethics Committee of Third Central Hospital, Tianjin.
Genomic DNA was extracted from tumor tissue samples by digesting with SDS/proteinase K in TE buffer followed by a standard phenol/chloroform extraction. The extracted DNA was subjected to bisulfite treatment as previously described[10]. Briefly, 1-2 μg genomic DNA was denatured with 0.3 mol/L NaOH at 37°C for 20 min, and incubated in 3.0 mol/L sodium bisulfite and 10 mmol/L hydroquinone at 55°C for 16 h. The DNA was desalted with a QIAquick gel extraction kit (Qiagen, Valencia, CA) and dissolved in 50 μL of 10 mmol/L TE buffer (pH 8.0). Then, 5.5 μL of 3.0 mol/L NaOH was added and incubated at 37°C for 20 min to desulfonate it. The modified DNA was neutralized with 30 μL of 10 mol/L ammonium acetate, precipitated using 2 volumes of ethanol, and resuspended in 40 μL of 1.0 mmol/L TE buffer (pH 7.6).
Fluorogenic quantitative MSP assay was carried out in the ABI Prism 7000 sequence detection system (Applied Biosystems, Foster City, CA). For each gene of the DKK family, a set of primers and a probe covering multiple CpG dinucleotides were designed within the putative CpG islands around the gene promoters. For the endogenous reference gene, ACTB (β-actin), the primers and probe were designed to avoid CpG dinucleotides. All primers and probes were designed according to the bisulfite-converted DNA sequences. Genes of interest were amplified in proportional to the degree of CpG methylation, while ACTB was amplified independently of its methylation. The sequences of primers and probes are as follows: (1) DKK1, 5'-GGGTTCGCGGTATAAAGGTAGTC-3' (sense), 5'-TCCGAAAACCCCCTACGATC-3' (antisense), 5'-FAM-TGGCGGTGGCGGCGTAGAGT-BHQ1-3' (Probe); (2) DKK2, 5'-GCGAGCGTAGCGTAAGTTCGT-3' (sense), 5'-CACTCACAATTACCCCGAAACG-3' (antisense), 5'-FAM-AGGTATCGTTGCGTTGGTAGCGATTCG-BHQ1-3' (Probe); (3) DKK3, 5'-GGTATCGGCGTTGTCGTATTTC- 3' (sense), 5'-CCACCCCGACTAAACCGAAT-3' (antisense), 5'-FAM-TCGCGGTTCGTTTATCGCGTC-BHQ1-3' (Probe); and (4) ACTB, 5'-TGGTGATGGAGGAGGTTTAGTAAGT-3' (sense), 5'-AACCAATAAAACCTACTCCTCCCTTAA-3' (antisense), 5'-FAM-ACCACCACCCAACACACAATAACAAACACA-BHQ1-3' (Probe).
Q-MSP assay was performed in a reaction volume of 20 μL in 96-well plates, and the final reaction mixture was consisted of 1 × real-time PCR master mix (Toyobo Co., Ltd. Shanghai), 200 nmol/L probe, 400 nmol/L primers of each gene, and 2 μL bisulfite-converted DNA templates. PCR was performed at 95°C for 2 min, followed by 45 cycles at 95°C for 15 s, and at 60°C for 45 s. Reactions were done in duplicate and each plate included at least 3 controls with no template (NTC), as well as negative and positive controls on each plate. Leukocyte DNA collected from a healthy individual was used as a negative control. The DNA methylated in vitro with SssI methyltransferase (New England Biolabs Inc., Beverly, MA) was used as a positive control for all studied genes.
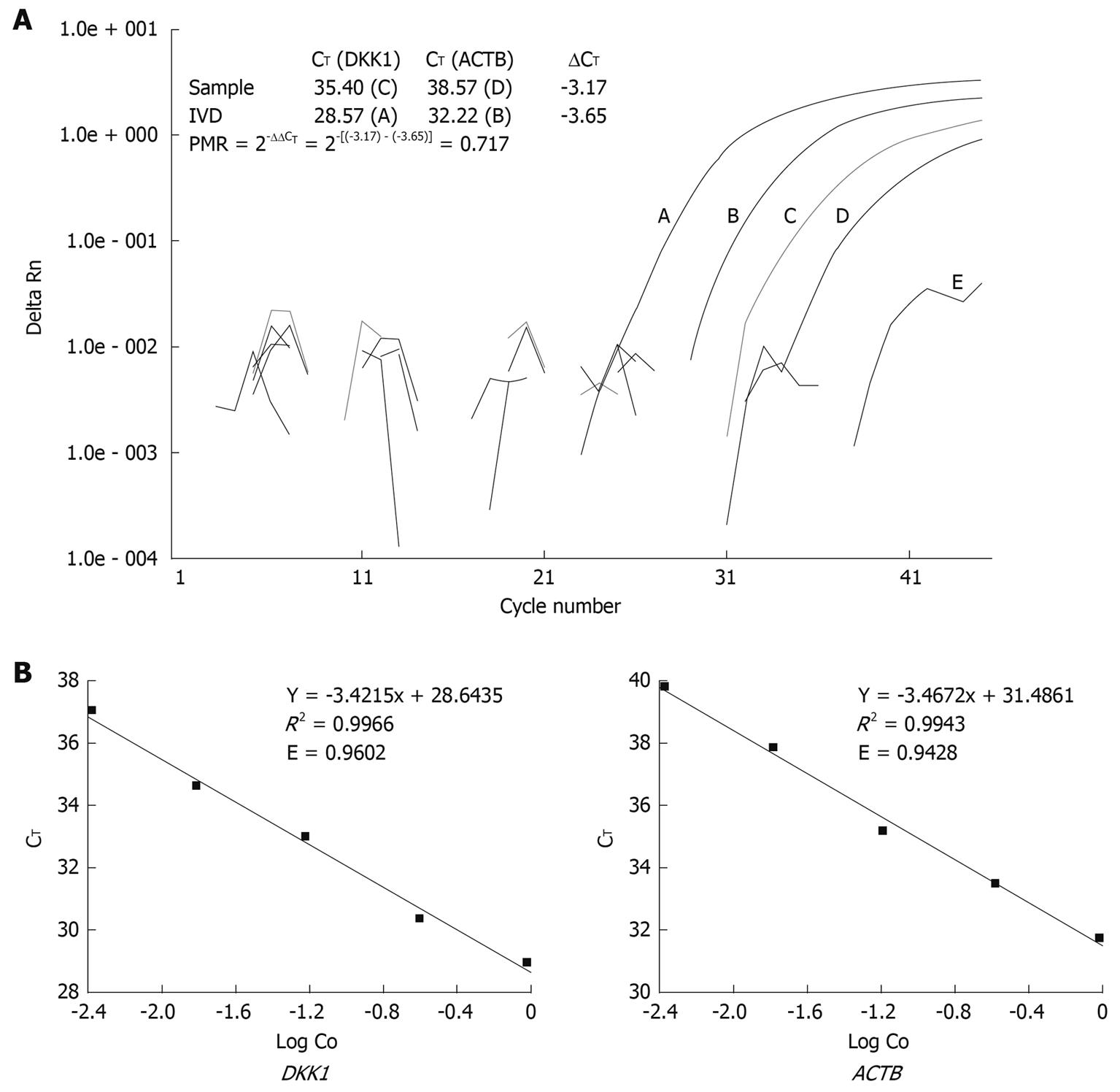
Abbreviation PMR was used to define the percentage of fully methylated molecules at a specific locus as previously described[11]. Briefly, the PMR value was calculated by dividing the GENE: ACTB ratio in a sample by the GENE: ACTB ratio in SssI-treated leukocyte DNA (IVD) and multiplied by 100. Parallel PCR reactions were done for the genes of interest and reference. Given the high efficiency of Q-MSP amplification for both DKK and ACTB genes in this study, PMR values were detected with the comparative CT method instead of the relative standard curve method, which needs serial dilutions of bisulfite-treated universally methylated DNA to construct a relative standard curve for each gene[12]. Relation between the percentages of methylated DNA molecules and CT was described as PMR = 2-ΔΔCT× 100% where ΔΔCT = ΔCT(Gene) - ΔCT(ACTB) = [CT(Gene) - CT(ACTB)](Sample) - [CT(Gene) - CT(ACTB)](IVD).
The number of cycles at which the fluorescence signal crossed a detection threshold was determined automatically with the ABI prism 7000 detection system, and referred to as CT. Representative Q-MSP amplification plots for DKK1 and corresponding calculation for PMR are illustrated in Figure 1A. For the ΔΔCT calculation to be valid, the efficiencies in target and reference amplification should be within 10%. PCR efficiency was calculated and compared according to the following equation recommended in technical manual of Applied Biosystems (Figure 1B).
RNA was extracted from HCC and matched NCL tissue samples using Trizol (Tiangen, Beijing) according to its manufacturer’s instructions. Total mRNA was digested with DNase I (Ambion, Austin, TX) to remove genomic DNA contamination and then subjected to reverse transcription using the reverse transcription system (Promega, Madison, WI). For the reverse-transcriptase PCR of DKK3, the sense primer (5'-ATCACCTGGGAGCTAGAGCCTGATG-3') and anti-sense primer (5'-ACCTCTCTGGGCAGCAGGGATCTC-3') were designed. PCR was done on the ABI Prism 7000 sequence detection system in combination with the SYBR green teal-time PCR master mix (Toyobo Co., Ltd, Shanghai). Melting curve analyses following amplification were performed to assure the product specificity. Relative expression of DKK3 mRNA was normalized to the housekeeping gene GAPDH in the same cDNA using the comparative CT method. Primer sequences for GAPDH are 5'-CTCATGACCACAGTCCATGCCATCACTG-3' (sense) and 5'-CATGAGGTCCACCACCCTGTTGCTGTA-3' (anti-sense).
ROC curves were plotted to assess the PMR values of DKK family as diagnosis biomarkers, and their discriminatory capacity was evaluated by calculating the area under the curve (AUC). Generally, a truly useless test has an AUC of 0.5, while a perfect test (one that has zero false positives and zero false negatives) has an AUC of 1.0. For each gene of DKK family, the PMR values in HCC tissue samples were considered patient results, while the values in matched NCL tissue samples were considered control results.
Relation between categorical variables was determined by Pearson χ2 test or Fisher’s exact test. Difference in median of PMR values between paired HCC and NCL tissue samples was detected by Wilcoxon matched pairs test, difference in HCC tissue and LC biopsy samples was revealed by Mann-Whitney U test. Variables associated with overall survival or progression-free survival rate were tested using Kaplan-Meier estimates and compared by log-rank test. Relative risk (RR) of DKK3 methylation-related death and other clinical variables were estimated from a univariate Cox proportional hazards model. Multivariate Cox models were also constructed to estimate the RR for DKK3 methylation with adjustments for potential confounding risk factors. All statistical tests were two-sided and P < 0.05 was considered statistically significant. Statistical analyses were performed with GraphPad Prism V5.0 (GraphPad Software, San Diego, CA) and SPSS V11.0 software for Windows (SPSS Inc., Chicago, IL), respectively.
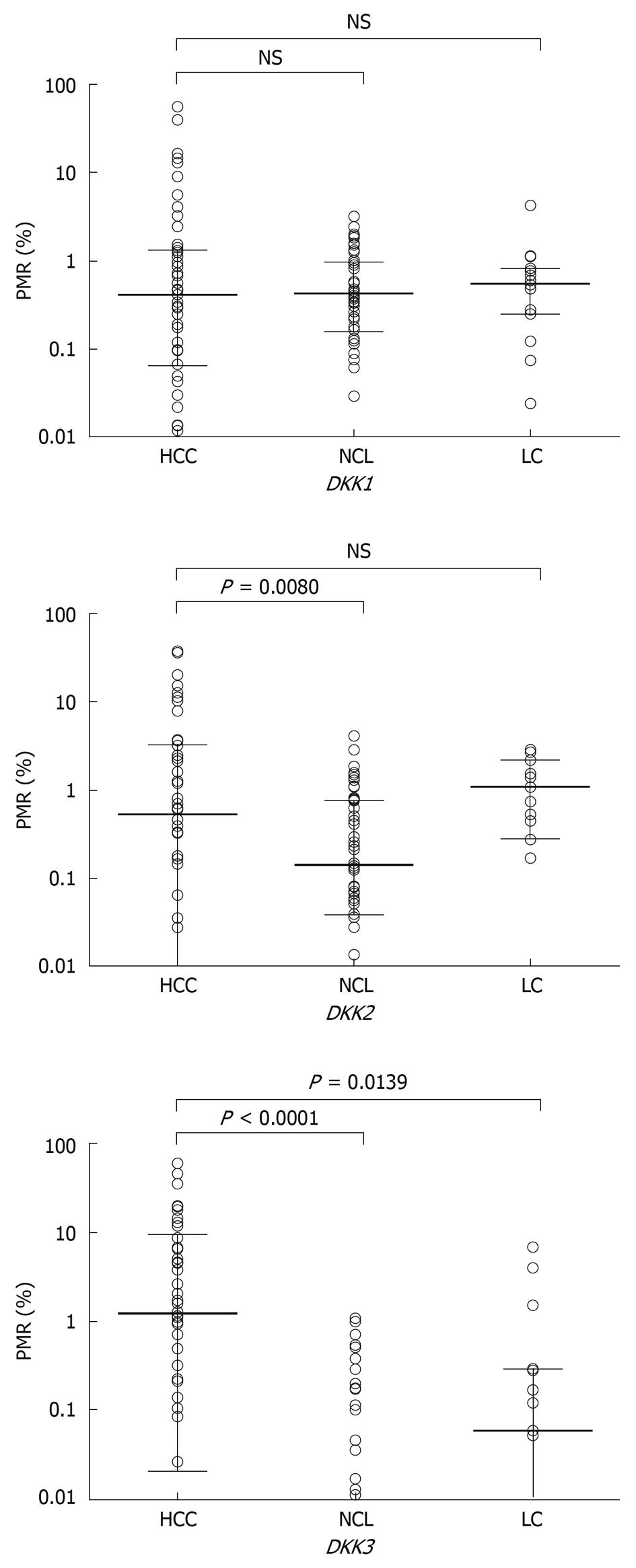
The methylation levels of DKK family in 3 kinds of tumor tissue sample were quantified by Q-MSP. The distribution of PMR values is illustrated in Figure 2. The methylation levels of DKK family in the 50 paired HCC tissue samples and NCL tissue samples were compared. Wilcoxon matched pairs test demonstrated that the methylation levels of DKK2 and DKK3 were significantly higher in HCC tissue samples than in corresponding NCL tissue samples (P = 0.0080, P < 0.0001), whereas no significant difference was found in the methylation level of DKK1. The difference in median PMR value was further compared between the 50 HCC and 15 LC tissue samples by Mann-Whitney test and a significant difference was only found in the median PMR value for DKK3 gene (P = 0.0139).
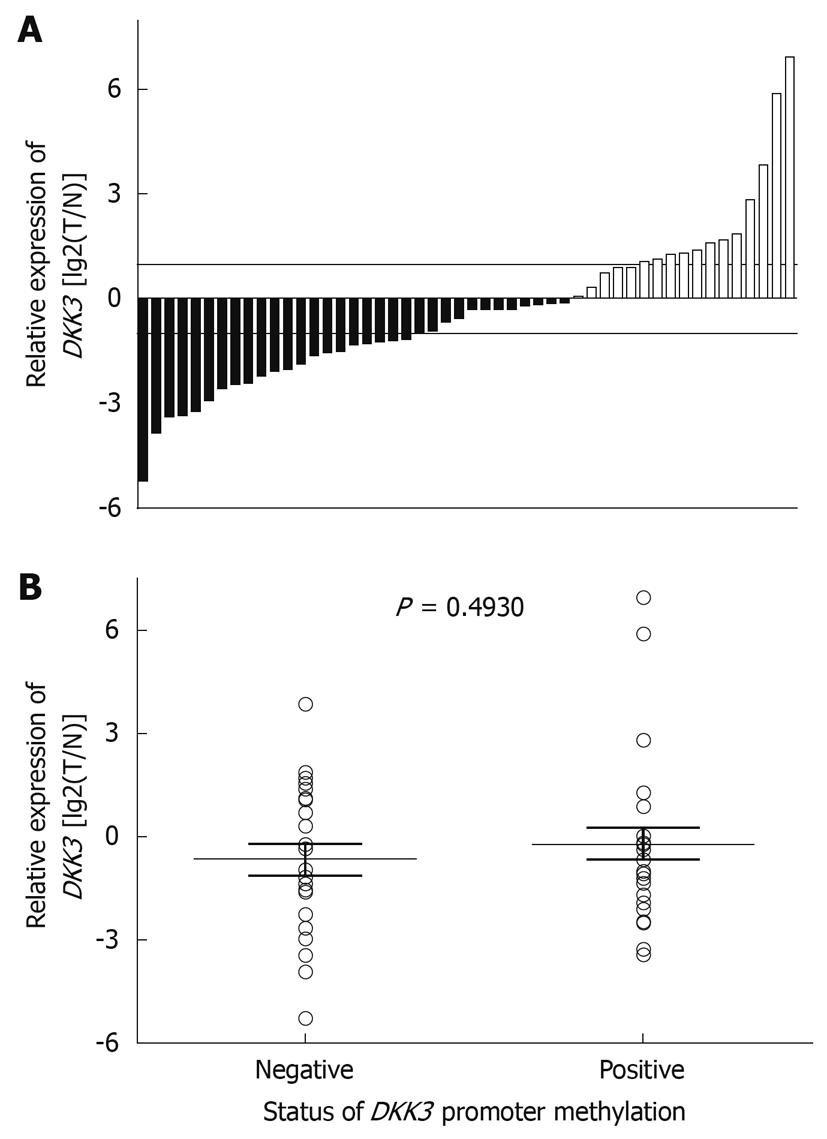
The expression of DKK3 mRNA in primary HCC tissue samples was detected by real-time PCR. Fifty pairs of HCC tissue samples and corresponding NCL tissue samples were analyzed. The expression level of DKK3 mRNA was lower in tumor tissue samples than in its adjacent tissue samples (Figure 3A). However, the median of DKK3 RNA expression was not statistically different between the methylated and unmethylated DKK3 genes (P = 0.4930) (Figure 3B).
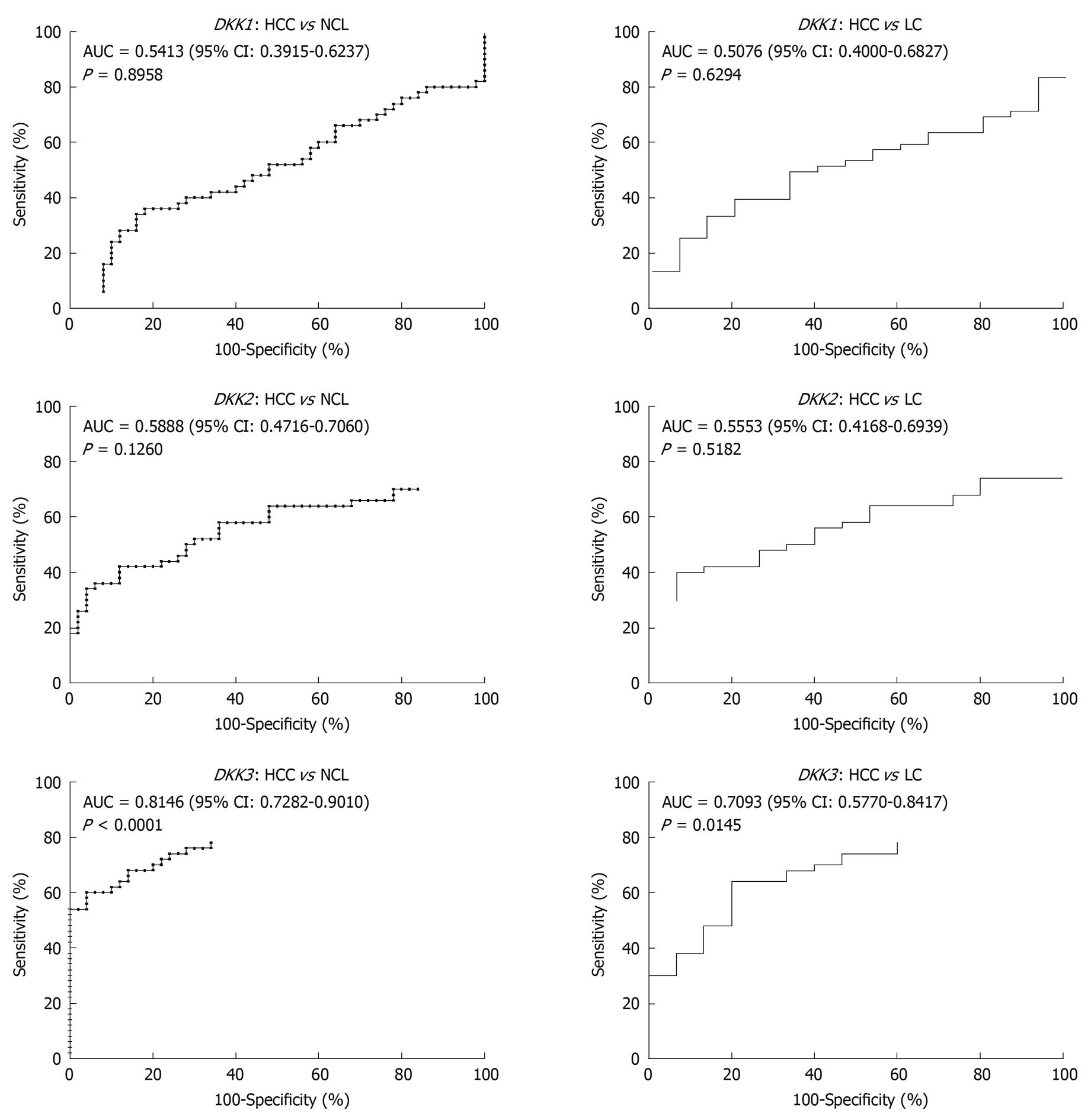
To assess whether quantitative methylation assay of DKK family can serve as a diagnosis tool to discriminate malignant from non-malignant liver tissue samples, ROC curves were plotted with PMR values as test results. The overall discriminatory ability of the test was evaluated by calculating the AUC (Figure 4). Two ROC curves were plotted for each gene. The PMR values in HCC tissue samples and the values in two types of liver cirrhosis tissue samples (NCL and LC) were considered patient results and control results, respectively. ROC curve analysis revealed that the AUC value for the PMR of DKK3 was relatively higher in the two kinds of tumor tissue samples (0.8146 for HCC vs NCL tissue samples and 0.7093 for HCC vs LC tissue samples, respectively).
To investigate the correlation between DKK family methylation and clinicopathological variables, the continuous PMR values were converted into discrete binary data, and the patients were also divided into two subgroups. To exclude the low DKK methylation level in a mere minority of cells, which may have little effect on gene activity in tumor tissue samples, PMR (1%) was selected as a criterion for the methylation of DKK. That is, the DKK methylation was classified into positive (PMR > 1%) and negative (PMR ≤ 1%) groups.
The methylation patterns of DKK genes in paired HCC and NCL tissue samples are summarized in Table 1. Consistent with the quantitative analysis above, significantly different methylation patterns of DKK3 were found in HCC and NCL tissue samples (P < 0.0001). The methylation frequency of DKK2 in HCC (64%, 32 of 50) and NCL (60%, 30 of 50) tissue samples was similar, while the methylation level of DKK2 was higher in HCC tissue samples (42%, 21 of 50) than in NCL tissue samples (16%, 8 of 50) (P = 0.0076). No significant difference of DKK1 methylation was found in HCC and NCL tissue samples.
| Positive (PMR > 1%) | Negative (PMR ≤ 1%) | P | |
| DKK1 | |||
| HCC | 14 | 36 | 0.6484 |
| NCL | 12 | 38 | |
| DKK2 | |||
| HCC | 21 | 29 | 0.0076 |
| NCL | 8 | 42 | |
| DKK3 | |||
| HCC | 27 | 23 | < 0.0001 |
| NCL | 2 | 48 |
Whether methylation of DKK family is related with certain clinicopathological variables was further determined. Statistical analysis showed that the methylation frequency of DKK1 or DKK2 was not related with the clinicopathological variables (data not shown). The frequency of DKK3 methylation was higher in multicentric HCC (Table 2).
| Positive (PMR > 1%) | Negative (PMR ≤ 1%) | P | |
| Total | 27 | 23 | |
| Sex | |||
| Male (n = 43) | 23 | 20 | |
| Female (n = 7) | 4 | 3 | 1.000 |
| Age (yr) | |||
| ≤ 55 (n = 28) | 14 | 14 | |
| > 55 (n = 22) | 13 | 9 | 0.5773 |
| Virus status | |||
| Positive (n = 44)1 | 22 | 22 | |
| Negative (n = 6) | 3 | 3 | 1.000 |
| AFP (ng/mL) | |||
| ≤ 400 (n = 30) | 17 | 13 | |
| > 400 (n = 20) | 11 | 9 | 1.000 |
| Tumor size (cm) | |||
| ≤ 5.0 (n = 19) | 9 | 10 | |
| > 5.0 (n = 31) | 18 | 13 | 0.5630 |
| Number of tumor | |||
| Single (n = 31) | 13 | 18 | |
| Multiple (n = 19) | 14 | 5 | 0.0417 |
| Portal vein invasion | |||
| Positive (n = 17) | 9 | 8 | |
| Negative (n = 33) | 18 | 15 | 1.000 |
To analyze the overall and progression-free survival rate associated with DKK3 methylation levels, patients were divided into methylation positive and negative groups according to their PMR values. The overall survival rate was not significantly different between the two groups, while the progression-free survival rate of patients with a high DKK3 methylation level was significantly lower than that of those with a low methylation level (P = 0.0255) (Figure 5). Univariate Cox proportional hazards model showed that the portal vein invasion and high DKK3 methylation level were related with an increased risk of disease progression when the tumor size was larger. Multivariate analysis model further showed that these three factors were the independently prognostic indicators for HCC (Table 3).
| Univariate analysis | Multivariate analysis1 | |||||
| RR | 95% CI | P | RR | 95% CI | P | |
| Tumor size (cm) | ||||||
| > 5.0 | 3.653 | 1.085-12.304 | 0.036 | 3.345 | 1.312-8.526 | 0.011 |
| ≤ 5.0 | 1 | 1 | ||||
| Portal vein invasion | ||||||
| Positive | 2.657 | 1.023-6.900 | 0.045 | 3.188 | 1.294-7.852 | 0.012 |
| Negative | 1 | 1 | ||||
| DKK3 methylation | ||||||
| Positive (PMR > 1%) | 2.370 | 0.965-5.823 | 0.060 | 2.527 | 1.063-6.008 | 0.036 |
| Negative (PMR ≤ 1%) | 1 | 1 | ||||
| AFP (ng/mL) | ||||||
| > 400 | 1.829 | 0.759-4.409 | 0.179 | - | ||
| ≤ 400 | 1 | |||||
| Age (yr) | ||||||
| > 55 | 1.552 | 0.569-4.234 | 0.391 | - | ||
| ≤ 55 | 1 | |||||
| Number of tumor | ||||||
| Multiple | 0.922 | 0.280-3.036 | 0.893 | - | ||
| Single | 1 | |||||
Development of HCC is a multistep process associated with genetic and epigenetic alterations. Methylation of multiple tumor suppressor genes is frequently observed in the development of cancer and may occur at different stages of HCC. However, not all these epigenetic alterations are directly involved in hepatocarcinogenesis. Aberrant methylation observed in HCC may be a consequence of the normal aging process, persistent viral infection, and chronic inflammation. Nishida et al[13] demonstrated that methylation of tumor suppressor genes in HCC is frequent but occurs in a gene-specific and disease-specific manner. Therefore, it is important to determine the prevalence and time of promoter hypermethylation in hepatocarcinogenesis, especially at the stage of hepatocellular transformation from a cirrhotic background. DKK1 acts as a powerful inhibitor of the Wnt signaling pathway, and epigenetic inactivation of DKK1 has been observed in various cancers[14-16]. In our study, a high frequency of DKK1 methylation was also observed in liver tissue samples (Table 1), whereas quantitative methylation analysis revealed that there was no statistically different DKK1 methylation in HCC and liver cirrhosis tissue samples, including tumor-adjacent cirrhosis and cirrhotic biopsy samples (Figure 2), suggesting that methylation of DKK1 may be involved in early hepatocarcinogenesis but not directly contributes to neoplasitc transformation from the liver cirrhotic background. Methylation of DKK2 has been observed in some kinds of tumor, but there is little direct evidence that epigenetic inactivation of DKK2 contributes to tumor development[14-16]. In our study, although DKK2 methylation occurred in tumor and corresponding cirrhosis tissue samples with a similar frequency (Table 1), the methylation level of DKK2 was obviously higher in tumor tissue samples than in its adjacent cirrhosis tissue samples (Figure 2), indicating that methylation of DKK2 may accumulate with the development of HCC. Interestingly, the DKK2 methylation level was higher in cirrhotic biopsy samples than in HCC tissue samples with no statistical significance, possibly due to the aberrant methylation in fibroblasts and stromal cells of cirrhotic biopsies[17].
In contrast to DKK1 and DKK2, DKK3 is methylated in a tumor-specific manner during hepatocarcinogenesis. In our study, DKK3 methylation occurred more frequently in HCC tissue samples than in cirrhosis tissue samples and the DKK3 methylation level was also dramatically higher in HCC tissue samples than in cirrhosis tissue samples, indicating that DKK3 promoter methylation may be an important event in the hepatocellular transformation from a cirrhotic background. Hsieh et al[18] found that DKK3 expression level is lower in human hepatoma tissue samples than in noncancerous liver tissue samples. It is thus reasonable to postulate that methylation and subsequently epigenetic inactivation of DKK3 gene may play an important role in hepatocarcinogenesis.
Although distinct methylation patterns of DKK3 were observed in HCC tissue samples and its adjacent liver cirrhosis tissue samples, no significant difference in mRNA expression was observed between the methylated and unmethylated groups. Similar results showing a lack of clear inverse correlation between the methylation and gene expression data have also been observed[19]. HCC tissue is very heterogeneous and our tissue samples were not microdissected to remove contaminated normal cells. The presence of a substantial amount of normal tissue in specimens prevents an exact assessment of the gene inactivation effects of CpG island hypermethylation. Gene expression in normal stromal and epithelial cells can mask a lack of expression in a subset of cells with CpG island hypermethylation[19]. Therefore, analysis of aberrant DNA hypermethylation is advantageous over gene expression analysis in that it has a greater sensitivity in the presence of contaminated normal cells[19].
The potential values of DKK gene methylation were further assessed in our study for clinical diagnosis purpose. The distinct methylation patterns of DKK gene (methylation frequencies or levels) in benign and malignant tissues are the prerequisite to determine a certain gene methylation as an effective molecular biomarker. Therefore, in addition to comparison of methylation frequencies in tumor and non-tumor groups, we further evaluated the discriminatory ability of quantitative methylation levels (PMR values) to distinguish HCC tissue from liver cirrhosis tissue using ROC analysis. Our results showed that the PMR values of DKK3 could discriminate HCC from liver cirrhosis with high sensitivity and specificity (Figure 4), suggesting that DKK3 methylation in combination with other diagnostic tools, may be a promising epigenetic biomarker for early detection of HCC.
It has been reported that aberrant DKK3 methylation is a major event in early and late liver malignant transformation and may constitute a critical target for risk assessment, treatment, and chemoprevention of HCC[20]. Therefore, another major question addressed in the present study concerns the prognostic value of DKK3 methylation in human HCC. In the present study, progression-free survival analysis showed that patients with DKK3 methylation tended to have relapse or metastasis shortly after resection (Figure 5). Multivariate Cox regression analysis further confirmed that DKK3 methylation was an independent prognostic indicator (Table 3). Interestingly, the number of tumors (single or multiple) was the only clinicopathological variable associated with DKK3 methylation in this study. HCC is prone to multicentric occurrence, and some other tumor suppressor genes are specifically methylated in multicentric HCC and can act as clonal markers[21]. Epigenetic inactivation of genes associated with multicentric occurrence may play an important role in the relapse or progression of HCC, thus patients with a high DKK3 methylation level can represent a subset of poor prognosis after surgical resection. Our results showed that a high DKK3 methylation level may serve as a potential prognostic factor for HCC. However, since the number of patients in the present study was relatively small, the prognostic significance of DKK3 methylation levels needs to be further investigated in a larger cohort of patients with a longer follow-up period.
In conclusion, methylation of DKK3 is an important event during early malignant transformation and progression of HCC, thus representing a prognostic indicator for risk assessment of HCC.
Aberrant promoter hypermethylation of tumor suppressor genes is very common in human cancers. It not only presents one of the important mechanisms in carcinogenesis, but also severs as a type of promising biomarkers for the diagnosis or prognosis of cancer patients.
Dickkopf (DKK) family is one class of the secreted Wnt antagonists. Its functional loss can contribute to activation of the Wnt pathway and result in carcinogenesis. Inactivation of Wnt antagonist genes and its epigenetic mechanism have been recently characterized in many cancers. However, few reports are available on the epigenetic silencing of DKK gene and its clinical significance in hepatocellular carcinoma (HCC).
Using quantitative methylation-specific polymerase chain reaction (Q-MSP) technology, the authors investigated the prevalence and time of DKK family methylation in adequate tumor tissue and biopsy samples. The results of this study demonstrate that methylation of DKK3 is an important event during early malignant transformation and progression of HCC, thus representing a prognostic indicator for risk assessment of HCC.
DKK3 methylation was identified as a specific epigenetic event involving hepatocellular transformation at cirrhosis stage, which not only provides a clue to the molecular basis of hepatocarcinogenesis, but also offers a potentially useful marker for the early diagnosis or prognosis of HCC.
Percent of methylated reference (PMR) is calculated by dividing the GENE/ACTB ratio in a sample by the GENE/ACTB ratio in SssI-treated DNA and multiplied by 100. The normalization performed to obtain PMR can simplify the cross-gene comparison, since the data range 0-100.
This is a very well-written paper trying to find anomalies in genes and their functions that may contribute to the prognostication and differential diagnosis of HCC with dysplastic nodules.
Peer reviewer: Dr. Paolo Del Poggio, Hepatology Unit, Department of Internal Medicine, Treviglio Hospital, Piazza Ospedale 1, Treviglio Bg 24047, Italy
S- Editor Tian L L- Editor Wang XL E- Editor Lin YP
| 1. | Fung SK, Lok AS. Management of patients with hepatitis B virus-induced cirrhosis. J Hepatol. 2005;42 Suppl:S54-S64. [Cited in This Article: ] |
| 2. | El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557-2576. [Cited in This Article: ] |
| 3. | Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, Thun MJ. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71-96. [Cited in This Article: ] |
| 4. | Rivenbark AG, Coleman WB. The use of epigenetic biomarkers for preclinical detection of hepatocellular carcinoma: potential for noninvasive screening of high-risk populations. Clin Cancer Res. 2007;13:2309-2312. [Cited in This Article: ] |
| 5. | Laurent-Puig P, Zucman-Rossi J. Genetics of hepatocellular tumors. Oncogene. 2006;25:3778-3786. [Cited in This Article: ] |
| 6. | Herath NI, Leggett BA, MacDonald GA. Review of genetic and epigenetic alterations in hepatocarcinogenesis. J Gastroenterol Hepatol. 2006;21:15-21. [Cited in This Article: ] |
| 7. | Niehrs C. Function and biological roles of the Dickkopf family of Wnt modulators. Oncogene. 2006;25:7469-7481. [Cited in This Article: ] |
| 8. | Zitt M, Untergasser G, Amberger A, Moser P, Stadlmann S, Zitt M, Müller HM, Mühlmann G, Perathoner A, Margreiter R. Dickkopf-3 as a new potential marker for neoangiogenesis in colorectal cancer: expression in cancer tissue and adjacent non-cancerous tissue. Dis Markers. 2008;24:101-109. [Cited in This Article: ] |
| 9. | Ding Z, Qian YB, Zhu LX, Xiong QR. Promoter methylation and mRNA expression of DKK-3 and WIF-1 in hepatocellular carcinoma. World J Gastroenterol. 2009;15:2595-2601. [Cited in This Article: ] |
| 10. | Lou C, Du Z, Yang B, Gao Y, Wang Y, Fang S. Aberrant DNA methylation profile of hepatocellular carcinoma and surgically resected margin. Cancer Sci. 2009;100:996-1004. [Cited in This Article: ] |
| 11. | Eads CA, Lord RV, Wickramasinghe K, Long TI, Kurumboor SK, Bernstein L, Peters JH, DeMeester SR, DeMeester TR, Skinner KA. Epigenetic patterns in the progression of esophageal adenocarcinoma. Cancer Res. 2001;61:3410-3418. [Cited in This Article: ] |
| 12. | Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101-1108. [Cited in This Article: ] |
| 13. | Nishida N, Nagasaka T, Nishimura T, Ikai I, Boland CR, Goel A. Aberrant methylation of multiple tumor suppressor genes in aging liver, chronic hepatitis, and hepatocellular carcinoma. Hepatology. 2008;47:908-918. [Cited in This Article: ] |
| 14. | Aguilera O, Fraga MF, Ballestar E, Paz MF, Herranz M, Espada J, García JM, Muñoz A, Esteller M, González-Sancho JM. Epigenetic inactivation of the Wnt antagonist DICKKOPF-1 (DKK-1) gene in human colorectal cancer. Oncogene. 2006;25:4116-4121. [Cited in This Article: ] |
| 15. | Lee J, Yoon YS, Chung JH. Epigenetic silencing of the WNT antagonist DICKKOPF-1 in cervical cancer cell lines. Gynecol Oncol. 2008;109:270-274. [Cited in This Article: ] |
| 16. | Suzuki R, Onizuka M, Kojima M, Shimada M, Fukagawa S, Tsuboi K, Kobayashi H, Shintani A, Ogawa Y, Kawada H. Preferential hypermethylation of the Dickkopf-1 promoter in core-binding factor leukaemia. Br J Haematol. 2007;138:624-631. [Cited in This Article: ] |
| 17. | Harder J, Opitz OG, Brabender J, Olschewski M, Blum HE, Nomoto S, Usadel H. Quantitative promoter methylation analysis of hepatocellular carcinoma, cirrhotic and normal liver. Int J Cancer. 2008;122:2800-2804. [Cited in This Article: ] |
| 18. | Hsieh SY, Hsieh PS, Chiu CT, Chen WY. Dickkopf-3/REIC functions as a suppressor gene of tumor growth. Oncogene. 2004;23:9183-9189. [Cited in This Article: ] |
| 19. | Eads CA, Lord RV, Kurumboor SK, Wickramasinghe K, Skinner ML, Long TI, Peters JH, DeMeester TR, Danenberg KD, Danenberg PV. Fields of aberrant CpG island hypermethylation in Barrett's esophagus and associated adenocarcinoma. Cancer Res. 2000;60:5021-5026. [Cited in This Article: ] |
| 20. | Calvisi DF, Ladu S, Gorden A, Farina M, Lee JS, Conner EA, Schroeder I, Factor VM, Thorgeirsson SS. Mechanistic and prognostic significance of aberrant methylation in the molecular pathogenesis of human hepatocellular carcinoma. J Clin Invest. 2007;117:2713-2722. [Cited in This Article: ] |
| 21. | Nomoto S, Kinoshita T, Kato K, Otani S, Kasuya H, Takeda S, Kanazumi N, Sugimoto H, Nakao A. Hypermethylation of multiple genes as clonal markers in multicentric hepatocellular carcinoma. Br J Cancer. 2007;97:1260-1265. [Cited in This Article: ] |