Published online Sep 7, 2010. doi: 10.3748/wjg.v16.i33.4135
Revised: April 16, 2010
Accepted: April 23, 2010
Published online: September 7, 2010
AIM: To design and validate broad-range 16S rRNA primers for use in high throughput sequencing to classify bacteria isolated from the human foregut microbiome.
METHODS: A foregut microbiome dataset was constructed using 16S rRNA gene sequences obtained from oral, esophageal, and gastric microbiomes produced by Sanger sequencing in previous studies represented by 219 bacterial species. Candidate primers evaluated were from the European rRNA database. To assess the effect of sequence length on accuracy of classification, 16S rRNA genes of various lengths were created by trimming the full length sequences. Sequences spanning various hypervariable regions were selected to simulate the amplicons that would be obtained using possible primer pairs. The sequences were compared with full length 16S rRNA genes for accuracy in taxonomic classification using online software at the Ribosomal Database Project (RDP). The universality of the primer set was evaluated using the RDP 16S rRNA database which is comprised of 433 306 16S rRNA genes, represented by 36 phyla.
RESULTS: Truncation to 100 nucleotides (nt) downstream from the position corresponding to base 28 in the Escherichia coli 16S rRNA gene caused misclassification of 87 (39.7%) of the 219 sequences, compared with misclassification of only 29 (13.2%) sequences with truncation to 350 nt. Among 350-nt sequence reads within various regions of the 16S rRNA gene, the reverse read of an amplicon generated using the 343F/798R primers had the least (8.2%) effect on classification. In comparison, truncation to 900 nt mimicking single pass Sanger reads misclassified 5.0% of the 219 sequences. The 343F/798R amplicon accurately assigned 91.8% of the 219 sequences at the species level. Weighted by abundance of the species in the esophageal dataset, the 343F/798R amplicon yielded similar classification accuracy without a significant loss in species coverage (92%). Modification of the 343F/798R primers to 347F/803R increased their universality among foregut species. Assuming that a typical polymerase chain reaction can tolerate 2 mismatches between a primer and a template, the modified 347F and 803R primers should be able to anneal 98% and 99.6% of all 16S rRNA genes in the RDP database.
CONCLUSION: 347F/803R is the most suitable pair of primers for classification of foregut 16S rRNA genes but also possess universality suitable for analyses of other complex microbiomes.
- Citation: Nossa CW, Oberdorf WE, Yang L, Aas JA, Paster BJ, DeSantis TZ, Brodie EL, Malamud D, Poles MA, Pei Z. Design of 16S rRNA gene primers for 454 pyrosequencing of the human foregut microbiome. World J Gastroenterol 2010; 16(33): 4135-4144
- URL: https://www.wjgnet.com/1007-9327/full/v16/i33/4135.htm
- DOI: https://dx.doi.org/10.3748/wjg.v16.i33.4135
Currently there is a broad international collaboration underway aimed at defining the microbiome (http://nihroadmap.nih.gov/hmp/) on the internal and external surfaces of the human body [Human Microbiome Project (HMP)]. The goals of the HMP collaborative project are to determine a core microbiome, assess how it changes during disease, and establish whether a change in the microbiome is associated with disease. It is anticipated that knowledge gained from the HMP will shed light on the etiology and pathogenesis of idiopathic chronic diseases that have a high impact on human health.
The human body is a highly complex conglomeration of human cells, bacteria, Archaea, fungi, and viruses, in which the number of microbes outnumbers the human cells by a factor of 10 to 1. Despite their intimate association, the microbial influence upon human development, physiology, immunity, and nutrition remains largely unstudied. This can be partly attributed to the lack of robust techniques needed for exploring a complex microbial community.
Previous attempts to fully characterize the human microbiome have had certain limitations. The classical method of culturing bacteria from human subjects excludes a large number of unculturable or not-yet-cultivated bacteria, and also misrepresents the abundance of some species due to selection by culture conditions. To overcome these drawbacks, culture-independent methods have been developed. The most commonly used culture-independent method relies on amplification and analysis of the 16S rRNA genes in a microbiome[1]. 16S rRNA genes are widely used for documentation of the evolutionary history and taxonomic assignment of individual organisms[2-6] because they have highly conserved regions for construction of universal primers and highly variable regions for identification of individual species[7]. Analysis of a microbiome is classically performed by amplifying all 16S rRNA genes in a sample, cloning the 16S amplicons into a vector transformed into Escherichia coli (E. coli), randomly selecting transformed colonies, producing high copies of the amplicon containing plasmid, purifying the plasmids, and sequencing the 16S rRNA inserts by the Sanger method[8].
Advantages of this protocol are potential identification of both cultivatable and uncultivable organisms and acceptable adequacy of single pass Sanger sequencing of 900-1000 bases for classifying bacteria. Disadvantages include annealing bias[9], cloning bias[10], as well as time and expenses. The high cost associated with this approach has been prohibitive for in-depth sampling of a complex microbiome. Inadequate sampling overlooks low abundance species. As a result, a low abundance species that could be a microbiome core species often cannot be consistently observed amongst different individuals. For example, a recent 16S rRNA gene survey of the human distal esophageal microbiome yielded 6800 sequences but revealed only one of the 166 species, Streptococcus mitis, shared by all 34 subjects sampled[11].
High throughput sequencing technology introduces a new way to perform microbiome surveys without limitations of cloning/Sanger sequencing. One 454 sequencing run can produce 1.2 million sequences in 8 h[12], which would require months or years of work with older methods. Because many sequences are acquired in one run, the unit cost per sequence read is a very small fraction of that for Sanger sequencing. This improvement allows sufficient sampling of a complex microbiome at affordable cost. The new technology also eliminates the cloning bias by directly sequencing the 16S rRNA genes generated by polymerase chain reaction (PCR). Therefore, high throughput sequencing is ideal if adaptable to meet the requirements needed for microbiome work.
The main limitation of high throughput sequencing is read length. Reads from next generation sequencing technologies are considerably shorter than those from Sanger sequencing. Illumina’s Solexa and Applied Biosystem’s SOLiD platforms generate reads of about 25-100 bases[13,14], while 454 sequencing technology reads up to 400-500 bases per sequence. The general approach to analyzing microbiomes in health and disease focuses at 2 levels. Population level analyses, such as the Fst test, P test, Unifrac analysis, clustering analysis, and normal reference range, relate samples of microbiomes by combined genetic distance between the samples. These types of analyses are relatively insensitive to variation in read length[15]. In Unifrac clustering analysis, reads of 100-200 nucleotides can yield the same clustering as full-length sequences if the correct regions are chosen for sequencing. Detailed analyses at the level of the operational taxonomic unit (OTU) depend on read length - the longer the more accurate[16]. Compared with full length sequences, single pass reads of approximate 900 bases (of full length 1500-1600) from Sanger sequencing is associated with a slight loss of classification accuracy, which has been acceptable considering the significant reduction in sequencing cost from sequencing the entire 16S rRNA genes.
In the recent studies utilizing 454 sequencing technology to perform 16S rRNA gene surveys of microbiomes[17-20], a major concern has been reduction of classification accuracy with short sequence reads. Several strategies have been tried to maximize the information obtained from short sequences. One is to utilize a paired-end sequencing strategy to increase sequence length[21]. Another is to target certain hypervariable regions (HVR) that are most informative for a specific microbiome of interest. Currently, various HVRs, individually[17,19] or in combination[18,22], have been used in analysis of a microbiome but their efficacies often are not validated.
Early studies using Sanger sequencing revealed interesting associations between human microbiomes and disease, such as that seen between the human foregut microbiome, dental/periodontal diseases and gastroesophageal reflux diseases (GERD)[11,23,24]. To further study such associations, the Human Microbiome Project[25] (part of the NIH Roadmap Initiative) currently supports in-depth analysis of the association between the human microbiome of various anatomical sites and related diseases. Our area of focus is the foregut microbiome during disease progression from GERD to esophageal adenocarcinoma (the fastest increasing cancer in the Western world). As the first step of the foregut microbiome project, we identified the most informative HVRs, and designed and validated a broad range primer set most suitable for the foregut microbiome. These will be used to facilitate our in-depth analysis of the human foregut microbiome and its role in GERD-related disease progression.
Sequences collected for our in silico analysis were obtained from separate, previously conducted 16S rRNA gene surveys of 3 foregut sites. Esophageal 16S sequences (6800) were obtained from research by Yang et al[11], oral 16S rRNA gene sequences (2458) were from research by Aas et al[26], and gastric 16S rRNA gene sequences (839) were from research by Bik et al[27], totaling 10 097 sequences. In addition, we removed sequences that were derived from patient gastric samples with Helicobacter pylori (300), chimera sequences (127), as well as sequences with more than 8 bases missing after E. coli position 27 (186). The final foregut dataset contained 9484 sequences (2373 oral, 6626 esophageal, 485 gastric). These sequences were represented by 220 species. Because 16S rRNA-based operational classification criterion does not have sufficient discriminatory power to differentiate between Streptococcus pseudopneumoniae and Streptococcus pneumoniae[28], as the two species differ by only 5 base pairs (bp) between their 16S rRNA genes corresponding to a 0.03% difference (16S-based operational threshold for separation between two species is 3% diversity), they were considered as one species in this study. As a result, 219 species were represented in the dataset.
The 219 16S rRNA gene sequences were aligned using the NAST alignment program[29]. The program was set to recognize sequences at least 1250 bases long with at least 75% identity. Because NAST may remove bases not found to be sufficiently homologous, and thus unalignable, several sequences were truncated after alignment, particularly at the 5’ end. The missing portion of these sequences was manually replaced after alignment with the corresponding region from the GenBank sequence of the same species so that all 16S sequences would be complete, beginning from position 28 of the E. coli 16S rRNA gene.
To simulate the sequence data that would be obtained using specific primer pairs, representative amplicons were constructed from full length 16S rRNA gene sequences. The criteria for design of the representative amplicons was based on 454 requirements. Amplicons could be no more than 600 bases in total (including primers and nucleotide barcode) for optimal emulsion PCR. Because on average 454 read lengths are approximately 400 bases, only the portion of the amplicon that would be sequenced was considered. For example, for a forward read amplicon of a total 500 base pairs with 50 bases of forward primer and nucleotide barcode, only the first 350 bases after the primer would be read, with the final 50 bases ignored. These amplicons were designed using 6 universal primer sets from the European Ribosome Database[30] as shown in Table 1.
| Primer | # bases | Sequence (5’ to 3’) | Species with identical match (%) |
| Forward primers | |||
| 8F | 20 | AGAGTTTGATCCTGGCTCAG | n/a1 |
| 343F | 15 | TACGGRAGGCAGCAG | 99.1 |
| 517F | 17 | GCCAGCAGCCGCGGTAA | 93.2 |
| 784F | 15 | AGGATTAGATACCCT | 90.0 |
| 917F | 16 | GAATTGACGGGGRCCC | 84.0 |
| 1099F | 16 | GYAACGAGCGCAACCC | 73.1 |
| Reverse primers | |||
| 534R | 18 | ATTACCGCGGCTGCTGGC | 91.8 |
| 798R | 15 | AGGGTATCTAATCCT | 90.0 |
| 926R | 20 | CCGTCAATTYYTTTRAGTTT | 83.0 |
| 1114R | 16 | GGGTTGCGCTCGTTRC | 74.9 |
| 1407R | 16 | GACGGGCGGTGTGTRC | 91.3 |
| 1541R | 20 | AAGGAGGTGATCCAGCCGCA | n/a1 |
The simulated amplicons were generated by trimming full length sequences of the 219 foregut species using the MEGA version 4 program (MEGA4)[31]. Aligned FASTA sequences were uploaded onto MEGA4, with the sequence file including an aligned E. coli 16S rRNA gene sequence as a positional template.
To simulate data that would be obtained by cloning/Sanger sequencing methods, amplicons of approximately 900 bases downstream of the starting position were first generated for the 219 sequences. These amplicons were used to theoretically compare how reads obtained by 454 sequencing technologies would compare to Sanger sequence reads. The 8F primer was located in the 219 aligned sequences (bases 8-27) and all bases upstream of 28 were deleted. The gaps from the aligned sequences were then removed, and all bases downstream of 928 were deleted leaving the sequences between positions 28 and 928 in E. coli (900 bases total) as a reference for the Sanger sequencing data that would be obtained for the 219 species.
For length-based amplicon comparison, aligned sequences were again trimmed upstream of E. coli position 28, representing the first base after the 8F forward primer. This E. coli position 28 was set as base #1. All gaps were then deleted from the aligned sequences and downstream bases were trimmed to leave amplicons with 900, 800, 700, 600, 500, 400, 300, 200, 100 and 50 bases. These were compared against full length 16S rRNA gene sequences for classification accuracy.
For amplicons based on different primer sets (within varying regions of the 16S rRNA gene), sequences of reverse and forward reads from both ends of the amplicons were analyzed. To trim full length sequences to the amplicons of interest, the position of the forward or reverse primer was first located by searching for the primer sequence. For theoretical forward reads of the amplicons, the sequence upstream and including the forward primer was deleted. Gaps were then deleted in the aligned sequences leaving all sequences with the base after the forward primer as base #1. From this position, the site of 350 bases downstream of position #1 was determined, and all bases downstream of #350 were deleted. This represented 454 sequencing’s ability to read the 350 bases downstream of the primer (after the first 50 or so bases of the primers and barcodes were read). For theoretical reverse reads of amplicons, the position of the reverse primer was located as before. From the 3’ end of the reverse primer complement (sense strand) the first base of the sequence of interest was designated position #1’. Using the E. coli 16S rRNA gene as a reference, the site of 360 bases upstream of base #1’ was determined and set as #360’. All bases upstream of #360’ were deleted. The gaps in the aligned sequences were then deleted and the number of bases in each sequence was determined. For any sequences with more than 350 bases, extra bases were removed manually from the 5’ end so that all sequences were 350 bases long. All sequence trims were then saved as FASTA files for analysis.
Sequence trims for size and designed amplicons were classified at the phylum to genus level using RDPII Classifier[32] and at the species level using RDPII Seqmatch[33].
For classification at the phylum to genus level, FASTA files were uploaded onto the RDPII Classifier at 80% confidence threshold and results were viewed at a display depth of 7 to see assignment data down to the genus level. The resultant classification dataset for the trimmed sequences was individually compared to the dataset obtained using full length sequences and/or 900 bp Sanger length sequences. The number of discrepant assignments was recorded.
For classification at the species level, FASTA files were uploaded onto RDPII Seqmatch. The following parameters were selected: typed and non typed strains, uncultured and isolates, ≥ 1200 bases, good quality, nomenclature taxonomy. The species classification of every sequence was individually analyzed and compared to the species that the full length version of the sequence was assigned to, as described previously[11].
Universal 16S rRNA primers were obtained from the European ribosomal RNA database[30]. The primer sequences are shown in Table 1. Determination of the percentage of the 219 foregut species with the exact primer sequence was done using MEGA4 software[31].
The optimized primers were searched using Probe Match against the bacterial 16S rRNA gene database at Ribosomal Database Project (http://rdp.cme.msu.edu).
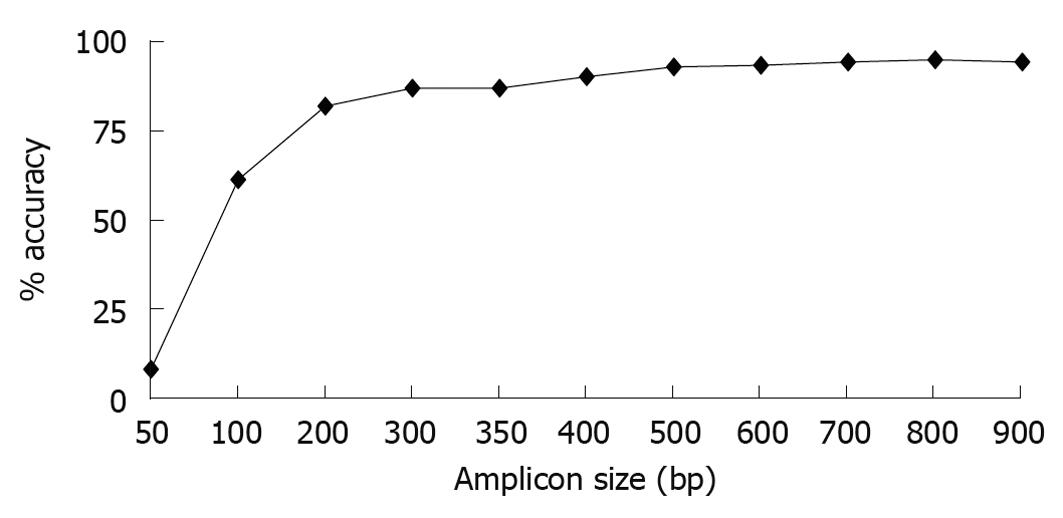
To compare shorter reads that would be obtained with high throughput sequencing against longer reads that would be obtained using Sanger sequencing, we performed in silico analysis; looking at the capability of different length 16S rRNA gene sequences to accurately classify foregut bacteria. Using full length sequences from 219 representative foregut species, we created sequence truncations that were 50-900 bases long beginning from base 28, the first base after the 8F primer. A length of 900 bases was chosen as the longest sequence to analyze because it is the length of a single pass Sanger sequence and generally accepted as accurate taxonomically. Each of the truncated versions of the 16S rRNA gene sequences was analyzed using RDPII classifier down to the genus level. The results showed that loss of classification accuracy was length dependent (Figure 1). At the genus level, sequences as small as 200 bases showed relatively good classification accuracies (94.1% accuracy for 900 base sequence, 90.0% for 400 bases, and 81.7% for 200 bases). Once the sequences became smaller than 200 bases, the classification accuracies decreased considerably, with the 100 base sequence having only 61.2% accuracy and the 50 base sequence having only 8.2% accuracy at the genus level. Overall, for identification at the genus level, the data showed that sequence sizes generated by Solexa or SOLiD platforms would have drastically lower classification accuracies (less than 50 bases, 8%-61% accuracy) than those sequence sizes generated by 454 technology (over 400 bases, over 90% accuracy).
The 16S rRNA gene contains 9 defined HVR[7]. These HVRs are of varying usefulness in classification depending on which species is being classified[22]. Therefore it would be necessary to choose the most informative region to sequence if less than 400 bases can be covered. To determine which region provided the most accurate classification data for the 219 foregut representative species, we undertook an analysis of different 350-bp regions of the 16S rRNA gene.
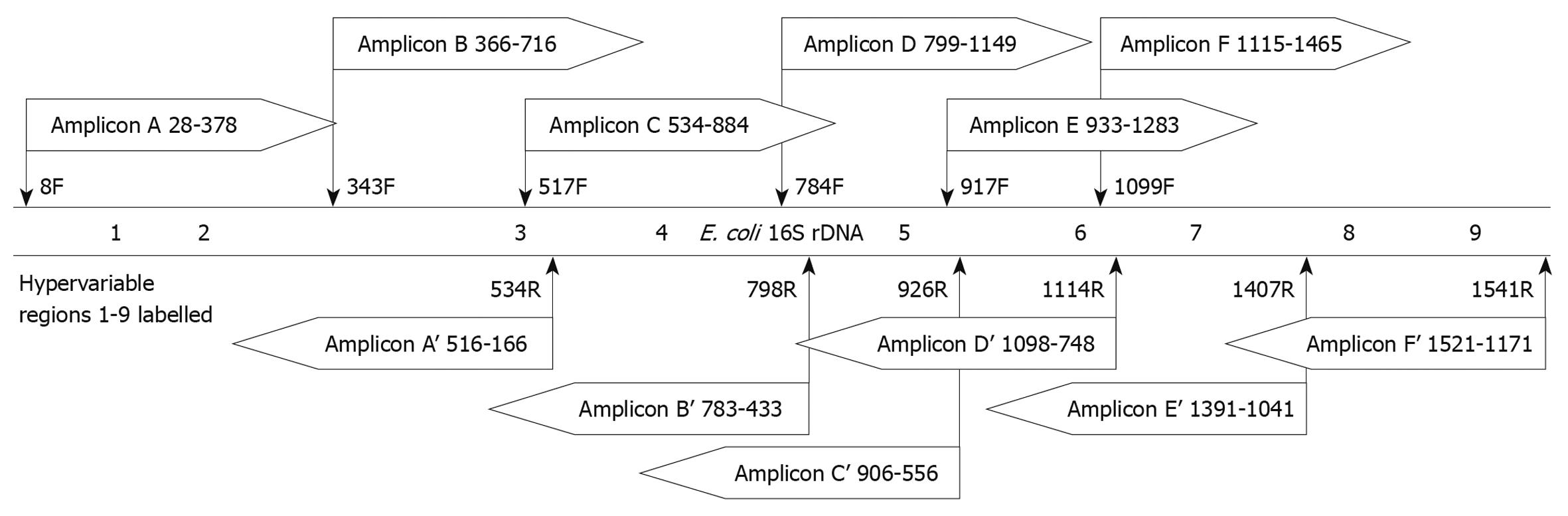
In the above size-based analysis, the 350-bp amplicon (determined to have 86.8% classification accuracy) (Figure 1) was located at the beginning of the 16S rRNA gene, starting immediately after the 8F primer, spanning bases 28-428 and including HVRs 1 and 2. To identify other potential 350-bp regions within the 16S rRNA gene that may have better classification accuracy, we analyzed 6 350-bp regions that simulated amplicons generated using various combinations of universal primers and covered the 9 HVRs (Table 2). Sequence reads from the designed amplicons were simulated by trimming the full length 16S rRNA gene sequences of the 219 foregut species as described in Materials and Methods. Because all amplicons were longer than the maximum read length of 454 technology, 2 sequence reads were simulated from the amplicons. A forward read, analyzing the first 350 bases from the 5’ end of the sense strand, and a reverse read analyzing the first 350 bases from the 3’ end. In total, 6 amplicons were created (A-F), and 12 reads were analyzed (forward reads A-F and reverse reads A’-F’). The location and direction of the reads from these amplicons is illustrated in Figure 2.
| Amplicon | Primers | Total length | HVR(s) included | |
| F | R | |||
| A | 8F | 534R | 527 | 1,2,3 |
| B | 343F | 798R | 456 | 3,4 |
| C | 517F | 926R | 410 | 3,4,5 |
| D | 784F | 1114R | 331 | 5,6 |
| E | 917F | 1407R | 491 | 6,7 |
| F | 1099F | 1541R | 443 | 7,8,9 |
| Amplicon | % accuracy compared to 900-bp amplicon/full length | ||||
| Phylum | Class | Order | Family | Genus | |
| Forward reads | |||||
| A | 97.7/97.7 | 96.3/95.9 | 95.9/95.4 | 93.2/94.5 | 87.7/86.8 |
| B | 99.1/99.1 | 98.2/96.8 | 97.7/96.3 | 97.3/95.9 | 91.8/89.0 |
| C | 99.5/99.5 | 98.2/96.8 | 97.7/96.8 | 97.3/95.9 | 90.4/88.1 |
| D | 98.6/99.1 | 98.6/98.2 | 97.3/97.3 | 95.9/96.3 | 88.1/86.8 |
| E | 98.2/98.6 | 97.7/98.2 | 95.4/95.9 | 92.2/93.6 | 85.4/84.9 |
| F | 97.7/98.2 | 96.3/97.3 | 95.0/95.4 | 91.8/93.6 | 83.6/84.5 |
| Reverse reads | |||||
| A’ | 98.6/98.6 | 97.3/97.7 | 97.3/97.7 | 95.0/96.3 | 90.0/90.9 |
| B’ | 99.5/99.5 | 98.2/96.8 | 97.7/96.8 | 97.7/96.3 | 93.6/91.8 |
| C’ | 99.5/99.5 | 98.2/96.8 | 97.7/96.8 | 97.3/95.9 | 91.3/90.0 |
| D’ | 99.5/100 | 98.6/99.5 | 96.8/98.2 | 94.5/96.3 | 87.2/89.5 |
| E’ | 98.2/98.6 | 96.8/98.2 | 94.5/96.3 | 91.3/94.1 | 83.6/87.2 |
| F’ | 98.2/98.6 | 96.8/98.2 | 95.0/96.4 | 92.2/95.0 | 82.6/85.8 |
Using the RDPII classifier, we evaluated the classification accuracy of the 12 reads at the phylum, class, order, family, and genus levels (Table 3). At the phylum level, classification accuracies for the 12 proposed reads ranged between 97.7% and 100% and at the genus level from 84.5% to 91.8%. Read B’ was the most accurate at the genus level, covering bases 433-783 including the HVR 3 and 4. Its accuracy vs the 900-bp Sanger mimics was 93.6%.
To determine the classification accuracy of sequence read B’ at the species level, we used the B’ region from the full length 219 foregut species sequences and assigned each B’ sequence to a species level operational taxonomic unit (SLOTU) using RDPII Seqmatch. The assigned SLOTUs using sequence read B’ were compared to the SLOTUs assigned with full length or 900 bp 16S rRNA gene sequences. At the species level 14 of the 219 foregut species were misclassified vs full length sequences (93.6% accuracy) (Table 4), compared to 10 of the 219 foregut species misclassified vs 900 bp sequences (95.4% accuracy) (data not shown).
| Species | Weight (of 9484) | Species identified using amplicon B |
| Atopobium AY959044 | 8 | Atopobium parvulum |
| Bacteroides vulgatus | 1 | Uncultured bacterium |
| Bradyrhizobium japonicum | 0 | Blastobacter denitrificans |
| Bradyrhizobium liaoningense | 1 | Blastobacter denitrificans |
| Escherichia fergusonii | 1 | Shigella sonnei |
| Escherichia flexneri | 4 | Shigella sonnei |
| Haemophilus aegyptius | 17 | Haemophilus influenzae |
| Haemophilus haemolyticus | 34 | Haemophilus |
| Lactobacillus gasseri | 2 | Lactobacillus johnsonii |
| Leptotrichia wadeii | 10 | Leptotrichia shahii |
| Neisseria macaca | 28 | Uncultured bacterium |
| Prevotella melaninogenica | 369 | Uncultured bacterium |
| Pseudoramibacter | 3 | Uncultured bacterium |
| Streptococcus infantis | 193 | Streptococcus mitis |
| Total | 671 |
It is important to note that our species level analyses of amplicon B’ was an unweighted analysis where each species was represented equally. Because not all of the 219 species are equally represented in the foregut, we used the results of the species level classification along with the experimentally determined relative abundance of each species in the foregut to give a weighted accuracy value. This provided a relative value of how many total sequences might be misclassified instead of how many species are misclassified. The relative abundance of each species in the foregut was based on the number of sequences of each species found in the foregut.
Of the 9484 sequences from the 3 studies, 671 would have been found to be classified inaccurately with amplicon B’, or approximately 7% giving a weighted species level classification accuracy of 92.9% for amplicon B’ (unweighted was 93.6%). Of those 671 sequences misclassified using sequence read B’, about half (369, or 55.0%) belonged to Prevotella melaninogenica and a significant number (193, or 28.8%) belonged to Streptococcus infantis, which was incorrectly identified as Streptococcus mitis.
Next, we directly examined the efficacy of amplicon B’ with every sequence from the esophageal study (n = 6800). The oral and gastric sequences were not analyzed because some were too short to span the full amplicon B’ region. We compared the taxonomical classifications obtained using the amplicon B’ truncations to those obtained using the original sequences (approximate 900 bp in length). Amplicon B’ accurately classified 6332 of the 6800 sequences (93.1% accuracy) at the species level. The 468 misclassified sequences belong to 14 species (Table 5).
| Species | Weight (of 6800) | Species identified by Amplicon B’ |
| Actinomyces naeslundii | 2 | Actinomyces viscosus |
| Atopobium AY959044 | 8 | Atopobium parvulum |
| Bacteroides vulgatus | 1 | Uncultured bacterium |
| Bradyrhizobium japonicum | 1 | Blastobacter denitrificans |
| Campylobacter showae | 4 | Campylobacter |
| Escherichia flexneri | 2 | Shigella sonnei |
| Haemophilus aegyptius | 17 | Haemophilus influenzae |
| Haemophilus haemolyticus | 33 | Haemophilus |
| Lactobacillus gasseri | 2 | Lactobacillus johnsonii |
| Leptotrichia wadeii | 7 | Leptotrichia shahii turn |
| Neisseria macaca | 25 | Uncultured bacterium |
| Prevotella melaninogenica | 288 | Uncultured bacterium |
| Pseudoramibacter | 1 | Uncultured bacterium |
| Streptococcus infantis | 77 | Streptococcus mitis |
| Total | 468 |
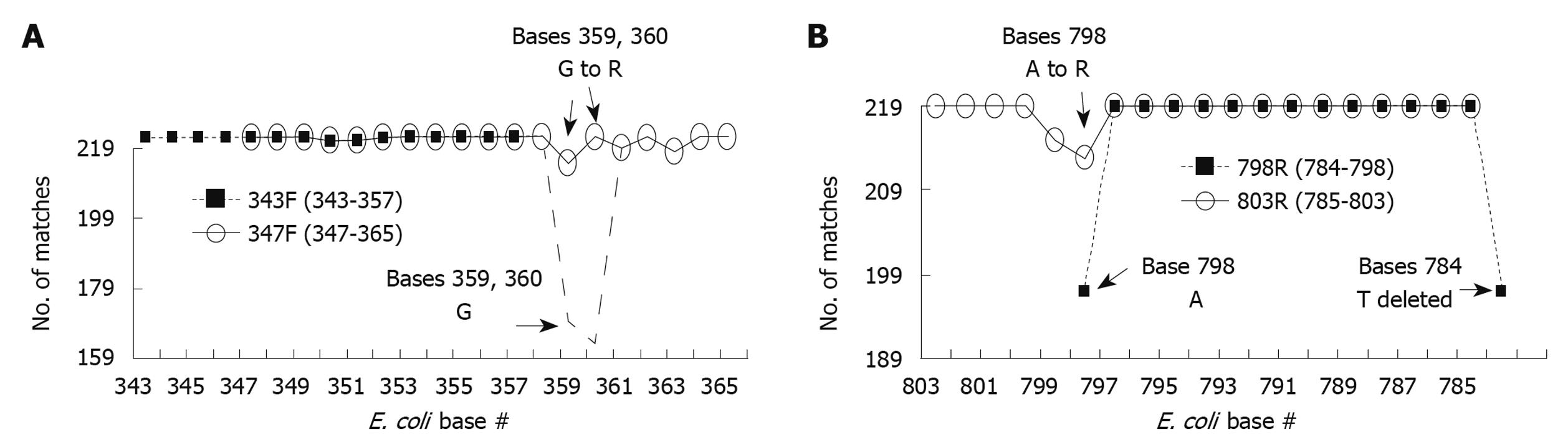
To maximize the probability of amplifying all bacterial species in the foregut, the 343F (15mer between positions 343 and 357) and 798R (15mer between positions 784 and 798) primer set were examined against 16S rRNA genes from these species to determine their universality. Of the 219 species, 217 (99.1%) had 100% homology to 343F and 197 (90.0%) had 100% homology to 798R. While the 343F primer had favorable homology with all sequences, it was extended 8 bases to position 365 and deleted 4 bases between positions 343 and 346 because the original primer sequence was too short, having a low melting temperature that would not have worked with PCR conditions when used in tandem with the 798R primer. This new primer, designated as 347F is a 19mer but this modification resulted in only 113 sequences (51.6%) with 100% homology. The 798R primer was also lengthened to base 803, but shortened from 784 to 785 to ensure CG at 5’ end of replication and to bring the melting temperature closer to the forward primer’s. This modified primer, designated as 803R is a 19mer and the modification resulted in only 180 sequences (81.4%) with 100% homology. To improve the universality of the modified 343F and 798R primers, we analyzed both at each individual base (Figure 3). For both the modified 343F and 798R there are positions of relatively low consensus. In the modified 343F, the consensus bases at positions 359 and 360 match with only 77.2% and 74.4% of the 219 species, respectively. In 798R, the consensus bases at positions 798 and 784 have 90.0% matches, respectively. To improve on these mismatches degenerative primers were constructed. Thus bases 359 and 360 in the modified 343F were changed from G (guanine) to R (guanine or adenosine). For 798R, base 798 was changed from A (adenosine) to R (guanine or adenosine) resulting in improved base matching as shown in Figure 3. The optimized 347F was 100% homologous with 205 (93.6%) species and 803R had 100% homology with 209 (95.4%) species. The sequences of the optimized primers are 5’-GGAGGCAGCAGTRRGGAAT (347F) and 5’-CTACCRGGGTATCTAATCC (803R).
The 347F/803R primer set has a broad taxonomic coverage of common foregut bacterial species identified from the limited number of sequences generated by Sanger sequencing, but deep sampling of the foregut microbiome by 454 sequencing will uncover numerous species that were below the detectable level by Sanger sequencing. To evaluate their potential to detect species not included in the foregut dataset, 347F/803R primer sequences were searched against the 16S rRNA gene database at RDP (http://rdp.cme.msu.edu/). The database has a collection of 16S rRNA genes from 5165 type strains and 433 306 high quality, near full length 16S rRNA genes from both cultured and uncultured bacterial species, representing 36 phyla. For optimized primer 347F, the introduction of ambiguity codons improved the coverage to 91.1% from 65.7% for the type strain sequences and to 90.4% from 63.7% for all 16S rRNA genes (Table 6). In comparison, the optimized primer 803R had an identical match with 91.8% of the type strain sequences and 84.9% of all 16S rRNA genes. Assuming a typical PCR can tolerate 2 mismatches between a primer and a template, the modified 374F will be able to anneal 99% of the type strain sequences and 98% of all 16S rRNA genes, compared with 99.9% and 99.6% for 803R primer, respectively.
| Primer | Optimization | Total species | Coverage at mismatches n (%) | Total sequence | Coverage at mismatches n (%) | ||||
| 0 | 1 | 2 | 0 | 1 | 2 | ||||
| 374F | Before | 5165 | 3392 (65.7) | 4835 (93.6) | 5042 (97.6) | 433 306 | 275 801 (63.7) | 406 626 (93.8) | 418 613 (96.6) |
| After | 5165 | 4703 (91.1) | 4996 (96.7) | 5114 (99.0) | 433 306 | 391 695 (90.4) | 418 832 (96.7) | 424 756 (98.0) | |
| 803R | Before | 5165 | 4584 (88.8) | 5091 (98.6) | 5159 (99.9) | 433 306 | 352 827 (81.4) | 417 612 (96.4) | 430 967 (99.5) |
| After | 5165 | 4741 (91.8) | 5131 (99.3) | 5159 (99.9) | 433 306 | 367 771 (84.9) | 427 791 (98.7) | 431 725 (99.6) | |
The introduction of next generation sequencing has been a boon to many fields of research, but has not yet been fully applied to microbial ecology. One reason for this is that previous read lengths of approximately 200 bases for 454 sequencing were not sufficient to accurately classify bacteria based on their 16S rRNA genes[33]. However, with the recent improvements to 454 sequencing which allows for longer (approximately 400 base) read lengths, this is now changing.
This increase in read length greatly improves 454’s applicability to 16S rRNA gene studies. We have shown that read lengths comparable to current 454 output (300-400 bases) give satisfactory 16S rRNA gene classification of bacteria at the genus and species level while shorter read lengths (such as those from Solexa and SOLiD technology) do not. However, with a change in sequencing read lengths of 16S rRNA from about 900 bp (as is commonly used with Sanger sequencing) to read lengths of 400, it is important to revisit which portion of the 16S rRNA gene to focus on to gain the most information from shorter reads. With the selection of the most informative stretch of the 16S rRNA gene, it is also important to ensure that the designed primers achieve a level of universality to be able to detect a wide array of microbial species. Although the primer pairs we chose to analyze were already available from the European Ribosomal Database, Wang has recently reported coverage rates of existing and predicted primers which span various regions of the 16S rRNA gene[34]. Some of those predicted primers with good coverage overlapped those that we had chosen.
Using the designed primers and 454 sequencing data, a complex microbiome can also be characterized by the relative abundance of 16S rRNA genes representing bacterial species in the microbiome, by assigning the 16S genes to specific species and calculating their relative weights. This is a much more accurate method of determining species relative abundance within the microbiome than the traditional methods such as colony forming unit counting (which is biased against fastidious bacteria) or cloning (which may be skewed due to cloning bias). As a comparison, the absolute amount of 16S rRNA genes from each organism can be determined by quantitative PCR (qPCR) but the number of species that can be tested in qPCR is limited.
The primer pairs we analyzed could theoretically be used for a wide range of bacteria-containing sources, such as environmental and clinical samples. By concentrating on species known to reside in the foregut (mouth, esophagus, and stomach), we have confirmed that 454 sequencing is an appropriate method for 16S rRNA gene analysis of the foregut microbiome. Careful choice of our primer set allowed us to find a region of the 16S rRNA gene that gives maximum classification accuracy within the current size limitations of 454 sequencing. This region was between the universal primers 343F and 798R, encompassing bases 361-784 and HVR 3 and 4. Focusing on just the 219 species of the foregut also allowed us to tailor our primers for better match, resulting in optimization of primers 343F and 798R (which we have modified to 347F and 803R).
We are using these primers in several foregut microbiome projects including one supported by the Human Microbiome Project. Without the time and cost restraints of cloning, 454 sequencing will allow us to analyze many more sequences than in previous foregut microbiome research (thousands vs 50-200 sequences per sample as was done previously). We expect that this more in-depth analysis of the foregut microbiome made possible by 454 sequencing will allow us to more clearly characterize the association between commensal bacteria of the esophagus and GERD-related esophageal disease progression to esophageal adenocarcinoma. In addition to identifying bacteria already shown to occupy the foregut, 454 sequencing will broaden the range of bacteria found within the human foregut with the increased coverage. This may lead to discovery of additional species, as well as previously undiscovered species, residing within the human foregut and a more accurate estimation of the species diversity of the foregut in normal and disease conditions.
Any species that was not included in our computational analyses will have a good chance of being identified by the foregut primers, based on our data showing the broad taxonomic coverage of these primers within domain bacteria. Thus, even though these primers were optimized for foregut species, they could potentially be used for other 16S rRNA based applications such as microbiome analysis on other anatomic sites or environmental samples. These carefully selected and designed primers will be essential tools in our efforts to harness the maximum potential that 454 sequencing offers to the field of microbial ecology of the human body.
The study of the human foregut microbiome has generated interest recently because of its association with gastroesophageal reflux disease-related complications. New technologies, such as 454 pyrosequencing, have increased the capability and scope of the study of complex microbiomes but have not yet been adapted for use with foregut microbiome samples.
16S rRNA gene analysis has previously been used to characterize the foregut microbiome. However, this was done with older techniques, such as Sanger sequencing, which did not allow for truly in-depth sequencing. In this study, the authors outline the size and location requirements for primer sets suitable to adapt 16S rRNA gene surveys to 454 pyrosequencing.
Previous 16S rRNA gene surveys used an amplicon of near full length, reading the first 900 or so bases. However, the restrictions of 454 sequencing limit amplicon size to 600 bases and read sizes to 400 bases. This is the first effort to systematically design a primer set to study the foregut microbiome using 454 pyrosequencing by testing different, suitably sized regions within the 16S gene for maximum accuracy of classification.
By designing a primer pair allowing accurate classification using 454 sequencing, this study will allow for in-depth characterization of the foregut microbiome, which is known to be associated with disease. Characterization of the disease-associated foregut microbiome may eventually lead to novel cures or diagnostics.
454 pyrosequencing is a next generation sequencing technology that allows for sequencing of DNA in a high throughput fashion (1 200 000 reads per run). 16S rRNA genes are ubiquitous, highly conserved markers that code for a ribosomal RNA unit. This gene marker is commonly used to identify bacteria at the species level.
The paper is well written and clearly illustrates its findings. The findings of this study are significant, as it provides a comparatively cheap and rapid way of identifying foregut bacteria. The study design and approach are sound. These results may replace the labor-intensive, time-consuming Sanger sequencing method. More importantly, this may lead to a detection method for foregut disease conditions.
Peer reviewers: Richard Anderson, PhD, Department of Anatomy and Cell Biology, University of Melbourne, Victoria 3010, Australia; Jong Park, PhD, MPH, MS, Associate Professor, Division of Cancer Prevention and Control, H. Lee Moffitt Cancer Center, College of Medicine, University of South Florida, 12902 Magnolia Dr. MRC209, Tampa, FL 33612, United States
S- Editor Wang JL L- Editor Cant MR E- Editor Lin YP
| 1. | Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697-703. [Cited in This Article: ] |
| 2. | Woese CR. Bacterial evolution. Microbiol Rev. 1987;51:221-271. [Cited in This Article: ] |
| 3. | Woese C. The universal ancestor. Proc Natl Acad Sci USA. 1998;95:6854-6859. [Cited in This Article: ] |
| 4. | Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci USA. 1990;87:4576-4579. [Cited in This Article: ] |
| 5. | Küntzel H, Heidrich M, Piechulla B. Phylogenetic tree derived from bacterial, cytosol and organelle 5S rRNA sequences. Nucleic Acids Res. 1981;9:1451-1461. [Cited in This Article: ] |
| 6. | Eigen M, Lindemann B, Winkler-Oswatitsch R, Clarke CH. Pattern analysis of 5S rRNA. Proc Natl Acad Sci USA. 1985;82:2437-2441. [Cited in This Article: ] |
| 7. | Van de Peer Y, Chapelle S, De Wachter R. A quantitative map of nucleotide substitution rates in bacterial rRNA. Nucleic Acids Res. 1996;24:3381-3391. [Cited in This Article: ] |
| 8. | Sanger F, Coulson AR. A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase. J Mol Biol. 1975;94:441-448. [Cited in This Article: ] |
| 9. | Suzuki MT, Giovannoni SJ. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl Environ Microbiol. 1996;62:625-630. [Cited in This Article: ] |
| 10. | Zoetendal EG, Akkermans AD, De Vos WM. Temperature gradient gel electrophoresis analysis of 16S rRNA from human fecal samples reveals stable and host-specific communities of active bacteria. Appl Environ Microbiol. 1998;64:3854-3859. [Cited in This Article: ] |
| 11. | Yang L, Lu X, Nossa CW, Francois F, Peek RM, Pei Z. Inflammation and intestinal metaplasia of the distal esophagus are associated with alterations in the microbiome. Gastroenterology. 2009;137:588-597. [Cited in This Article: ] |
| 12. | Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376-380. [Cited in This Article: ] |
| 13. | http://www.illumina.com/downloads/DeNovoAssembly_TechNote.pdf. [Cited in This Article: ] |
| 14. | http://www.appliedbiosystems.com/cms/groups/mcb_marketing/documents/general documents/cms_057562.pdf. [Cited in This Article: ] |
| 15. | Liu Z, Lozupone C, Hamady M, Bushman FD, Knight R. Short pyrosequencing reads suffice for accurate microbial community analysis. Nucleic Acids Res. 2007;35:e120. [Cited in This Article: ] |
| 16. | Wommack KE, Bhavsar J, Ravel J. Metagenomics: read length matters. Appl Environ Microbiol. 2008;74:1453-1463. [Cited in This Article: ] |
| 17. | Spear GT, Sikaroodi M, Zariffard MR, Landay AL, French AL, Gillevet PM. Comparison of the diversity of the vaginal microbiota in HIV-infected and HIV-uninfected women with or without bacterial vaginosis. J Infect Dis. 2008;198:1131-1140. [Cited in This Article: ] |
| 18. | Dowd SE, Sun Y, Secor PR, Rhoads DD, Wolcott BM, James GA, Wolcott RD. Survey of bacterial diversity in chronic wounds using pyrosequencing, DGGE, and full ribosome shotgun sequencing. BMC Microbiol. 2008;8:43. [Cited in This Article: ] |
| 19. | Andersson AF, Lindberg M, Jakobsson H, Bäckhed F, Nyrén P, Engstrand L. Comparative analysis of human gut microbiota by barcoded pyrosequencing. PLoS One. 2008;3:e2836. [Cited in This Article: ] |
| 20. | Roesch LF, Fulthorpe RR, Riva A, Casella G, Hadwin AK, Kent AD, Daroub SH, Camargo FA, Farmerie WG, Triplett EW. Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J. 2007;1:283-290. [Cited in This Article: ] |
| 21. | Fullwood MJ, Wei CL, Liu ET, Ruan Y. Next-generation DNA sequencing of paired-end tags (PET) for transcriptome and genome analyses. Genome Res. 2009;19:521-532. [Cited in This Article: ] |
| 22. | Chakravorty S, Helb D, Burday M, Connell N, Alland D. A detailed analysis of 16S ribosomal RNA gene segments for the diagnosis of pathogenic bacteria. J Microbiol Methods. 2007;69:330-339. [Cited in This Article: ] |
| 23. | Pei Z, Bini EJ, Yang L, Zhou M, Francois F, Blaser MJ. Bacterial biota in the human distal esophagus. Proc Natl Acad Sci USA. 2004;101:4250-4255. [Cited in This Article: ] |
| 24. | Pei Z, Yang L, Peek RM, Jr Levine SM, Pride DT, Blaser MJ. Bacterial biota in reflux esophagitis and Barrett's esophagus. World J Gastroenterol. 2005;11:7277-7283. [Cited in This Article: ] |
| 25. | Peterson J, Garges S, Giovanni M, McInnes P, Wang L, Schloss JA, Bonazzi V, McEwen JE, Wetterstrand KA, Deal C. The NIH Human Microbiome Project. Genome Res. 2009;19:2317-2323. [Cited in This Article: ] |
| 26. | Aas JA, Paster BJ, Stokes LN, Olsen I, Dewhirst FE. Defining the normal bacterial flora of the oral cavity. J Clin Microbiol. 2005;43:5721-5732. [Cited in This Article: ] |
| 27. | Bik EM, Eckburg PB, Gill SR, Nelson KE, Purdom EA, Francois F, Perez-Perez G, Blaser MJ, Relman DA. Molecular analysis of the bacterial microbiota in the human stomach. Proc Natl Acad Sci USA. 2006;103:732-737. [Cited in This Article: ] |
| 28. | Arbique JC, Poyart C, Trieu-Cuot P, Quesne G, Carvalho Mda G, Steigerwalt AG, Morey RE, Jackson D, Davidson RJ, Facklam RR. Accuracy of phenotypic and genotypic testing for identification of Streptococcus pneumoniae and description of Streptococcus pseudopneumoniae sp. nov. J Clin Microbiol. 2004;42:4686-4696. [Cited in This Article: ] |
| 29. | DeSantis TZ Jr, Hugenholtz P, Keller K, Brodie EL, Larsen N, Piceno YM, Phan R, Andersen GL. NAST: a multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucleic Acids Res. 2006;34:W394-W399. [Cited in This Article: ] |
| 30. | Wuyts J, Perrière G, Van De Peer Y. The European ribosomal RNA database. Nucleic Acids Res. 2004;32:D101-D103. [Cited in This Article: ] |
| 31. | Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596-1599. [Cited in This Article: ] |
| 32. | Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261-5267. [Cited in This Article: ] |
| 33. | Cole JR, Chai B, Farris RJ, Wang Q, Kulam-Syed-Mohideen AS, McGarrell DM, Bandela AM, Cardenas E, Garrity GM, Tiedje JM. The ribosomal database project (RDP-II): introducing myRDP space and quality controlled public data. Nucleic Acids Res. 2007;35:D169-D172. [Cited in This Article: ] |