Copyright
©2010 Baishideng.
World J Gastroenterol. Aug 21, 2010; 16(31): 3936-3943
Published online Aug 21, 2010. doi: 10.3748/wjg.v16.i31.3936
Published online Aug 21, 2010. doi: 10.3748/wjg.v16.i31.3936
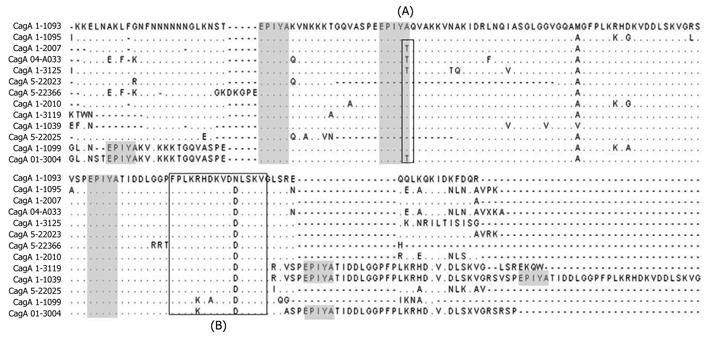
Figure 2 Multiple alignment of the amino acid sequence of the 3’ region from cagA.
First Square (A) shows the Glutamine acid-Proline-Isoleucine-Tyrosine-Alanine (EPIYA) motifs which had a modified EPIYA-B motif (EPIYT) instead of EPIYA. Second Square (B) shows the CagA multimerization (CM) motif which contains the amino acid sequence FPLXRXXXVXDLSKVG. The shaded squares represent the localization of EPIYA motif in the alignment. Amino acids are designated by standard one-letter codes. (∙) same residue; (-) gap. GenBank accession numbers and EPIYA motif of each isolate in this figure: CagA 1-1093 (FJ915841) (ABC), CagA 1-1095 (FJ915842) (ABC), CagA 1-2007 (FJ915843) (ABC), CagA 04-A033 (FJ915904) (ABC), CagA 1-3125 (FJ915893) (ABC), CagA 5-22023 (FJ915937) (AC), CagA 5-22366 (FJ915913) (ABC), CagA 1-2010 (FJ915844) (ABC), CagA 1-3119 (FJ915891) (ABCC), CagA 1-1039 (FJ915944) (ABCCC), CagA 5-22025 (FJ915878) (BC), CagA 1-1099 (FJ915912) (AABC) and CagA 01-3004 (FJ915917) (AABCC).
-
Citation: Acosta N, Quiroga A, Delgado P, Bravo MM, Jaramillo C.
Helicobacter pylori CagA protein polymorphisms and their lack of association with pathogenesis. World J Gastroenterol 2010; 16(31): 3936-3943 - URL: https://www.wjgnet.com/1007-9327/full/v16/i31/3936.htm
- DOI: https://dx.doi.org/10.3748/wjg.v16.i31.3936