Copyright
©2010 Baishideng.
World J Gastroenterol. Jan 21, 2010; 16(3): 289-295
Published online Jan 21, 2010. doi: 10.3748/wjg.v16.i3.289
Published online Jan 21, 2010. doi: 10.3748/wjg.v16.i3.289
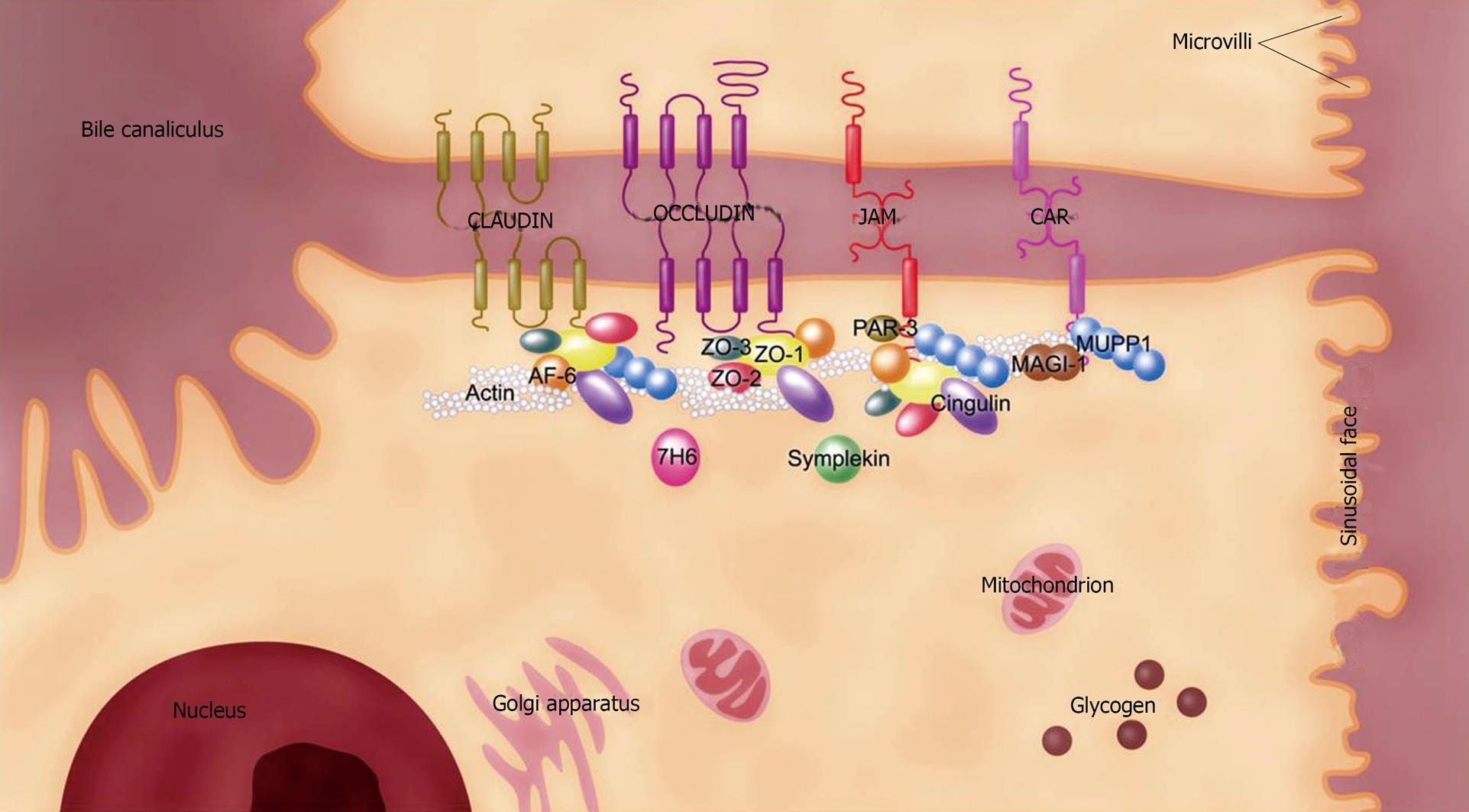
Figure 1 Molecular structure of the TJ in the mammalian liver.
The TJ in the liver is associated with hepatocytes and bile duct cells. TJ location around the bile canaliculus between 2 adjacent hepatocytes is shown. For simplicity, the molecular structure depicted in this figure represents the TJ molecules found in the mammalian liver. Claudin, occludin, JAM, and CAR are 4 core units for constituting TJ by uniting a panel of peripheral proteins like ZO-1 to form multiprotein complexes. TJ molecules display differential localizations in the mammalian liver, such that some of them like human symplekin and mouse CAR-2 are associated with both hepatocytes and bile duct cells while others, such as mouse CAR-1, are only found in the latter cell type. CAR: Coxsackievirus and adenovirus receptor; JAM: Junctional adhesion molecule; MAGI-1: Membrane-associated guanylate kinase inverted-1; MUPP1: Multiple PDZ domain protein-1; PAR-3: Partitioning defective 3 homolog; TJ: Tight junction; ZO: Zonula occludens.
- Citation: Lee NP, Luk JM. Hepatic tight junctions: From viral entry to cancer metastasis. World J Gastroenterol 2010; 16(3): 289-295
- URL: https://www.wjgnet.com/1007-9327/full/v16/i3/289.htm
- DOI: https://dx.doi.org/10.3748/wjg.v16.i3.289