INTRODUCTION
Gastric cancer remains one of the most common forms of cancer worldwide with approximately 870 000 new cases and 650 000 deaths per year[1–4], which accounts for about 9.9% of new cancers[5]. Worldwide, there has been a decline in the incidence of the intestinal type gastric cancer in the last few decades, following the overall decline in the incidence of gastric cancer. By contrast, the decline in the diffuse type gastric cancer has been more gradual. Some studies have reported an increase in the diffuse type of gastric carcinoma, especially the signet-ring cell type[6]. As a result, the diffuse type now accounts for about 30% of gastric carcinomas in some reported series[7]. Intestinal-type and diffuse-type gastric cancer differ in their epidemiology, pathogenesis, genetic profile, and clinical outcome[8].
In 1999, we cloned a gene segment GCRG123, which was down-regulated in gastric adenocarcinoma. BLASTX analysis revealed that the predicted GCRG123 product was a lamin-like protein[9]. Recently, we have used the updated GeneBank database for further analysis on the GCRG123 sequence. We found that GCRG123 appeared to be a long interspersed nucleotide element-1 (LINE-1, L1) family member. Additionally, we revealed that GCRG123 was ultimately up-regulated in stomach signet-ring cell carcinoma, as well as in normal pyloric glands and epithelia, which shows an opposite expression pattern compared with its down-regulation in stomach intestinal-type adenocarcinoma.
MATERIALS AND METHODS
Patients and tissue acquisition
All samples were obtained from the Department of Pathology, General Hospital of Chinese PLA. Specimens of paraffin-embedded gastric tissues, including 15 cases of gastric signet-ring cell carcinoma, 15 of advanced gastric intestinal-type adenocarcinoma and 15 of normal gastric mucosal tissues, were collected for in situ hybridization analysis. One set of fresh gastric signet-ring cell carcinomas and paired non-cancerous gastric tissues, and one set of fresh gastric intestinal-type adenocarcinomas and paired non-cancerous gastric tissues were used for Northern blot analysis. The diagnosis of cancer was confirmed through histology.
In situ hybridization
cRNA probe labeling: Digoxigenin-labeled anti-sense and sense cRNA probes were prepared by in vitro transcription (DIG RNA Labeling Kit (SP6/T7); Roche Diagnostics, Mannheim, Germany). Briefly, the following labeling procedure was employed: purified GCRG123 cDNA 100 ng/10 &mgr;L, 5 × NTP labeling mixture 4 &mgr;L, 5 × transcription buffer 4 &mgr;L, and RNA polymerase SP6/T7 2 &mgr;L were mixed gently, centrifuged, and then incubated for 1 h at 42°C. Two microliters of RNase-free DNase I was added to remove template DNA, by incubating for 15 min at 37°C, and the reaction was finally stopped by adding 2 &mgr;L 0.2 mol/L EDTA (pH 8.0). Labeling efficiency was directly detected by a spot test as described in the protocol of the kit.
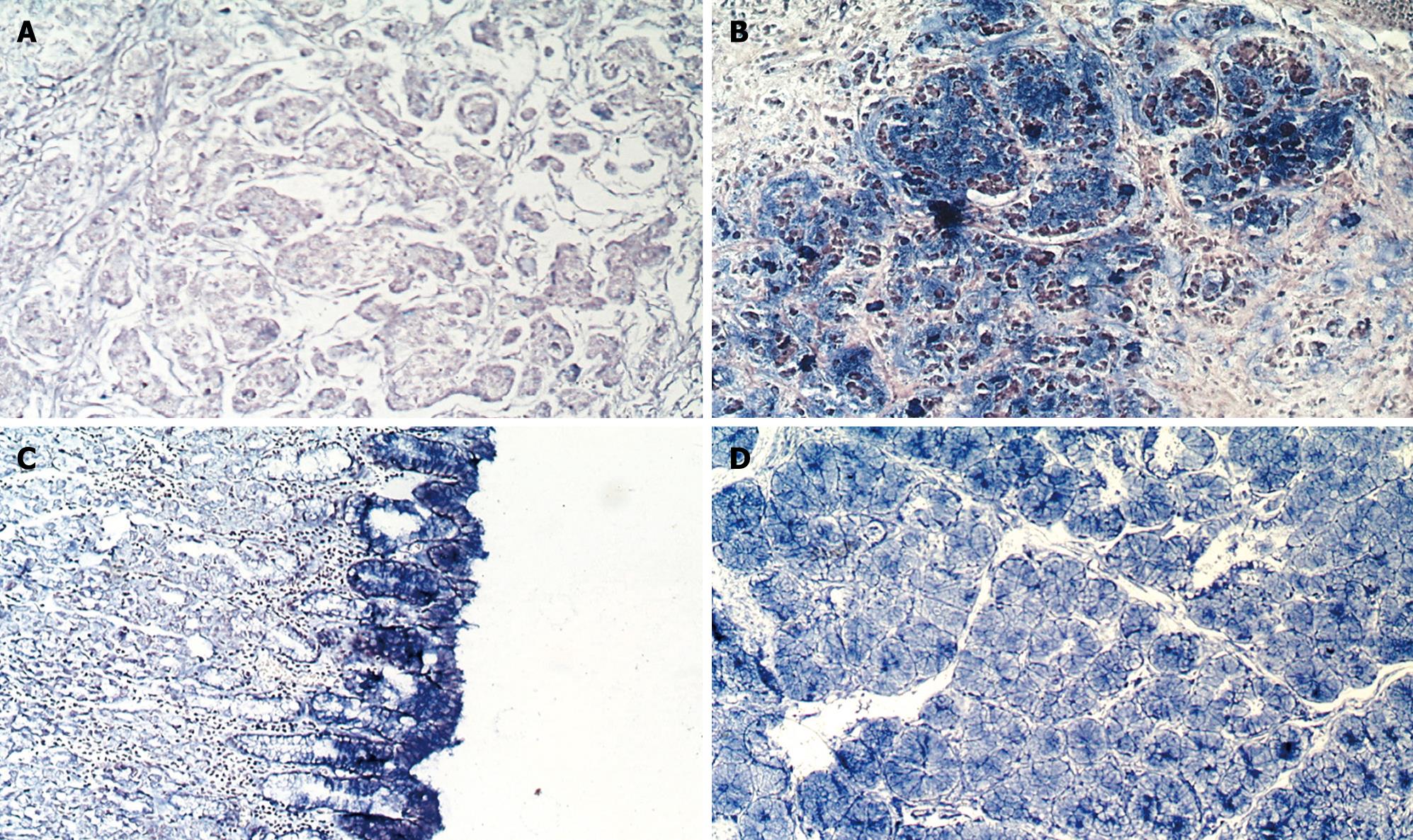
Hybridization: All specimens were fixed in 10% neutral-buffered formalin and embedded in paraffin. A series of 5-&mgr;m thick sections were cut for analysis. In situ hybridization was performed as previously described[1011] with some amendments, using digoxigenin-labeled anti-sense cRNA probes. Briefly, the slides were dried at 40°C overnight, dewaxed, rehydrated and pretreated with DEPC-treated PBS containing 100 mmol/L glycine and 0.3% Triton X-100, respectively. The sections were then permeabilized with 20 &mgr;g/mL RNase-free proteinase K (Merck, Darmstadt, Germany) for 20 min, incubated at 37°C for at least 20 min with prehybridization buffer. Each section was overlaid with 30 &mgr;L hybridization buffer containing 10 ng digoxigenin-labeled cRNA probe and incubated at 42°C overnight. After hybridization, the section was incubated with digoxigenin antibody (75 mU/mL) for 2 h. The positive signal for GCRG123 mRNA was detected by using NBT/BCIP (Promega, WI, USA) as a substrate. Sense cRNA probes were used as a negative control. The presentation of blue staining in the cytoplasm was considered positive. The positive staining of cytoplasm was scored manually as described below: -, barely detectable light blue; 1+, diffuse light blue; 2+, blue; 3+, dark blue. More than 100 non-tumor or tumor cells were quantified in each measurement, and more than one measurement was required to confirm the diagnosis. Consequent slides with H&E staining were then reviewed to compare the histological patterns to the staining patterns in the in situ hybridization preparations.
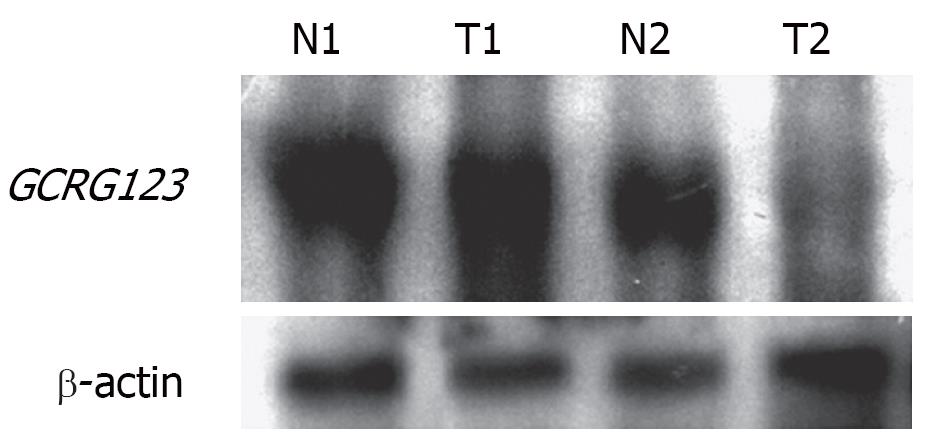
Northern blot analysis
Dig Northern Starter Kit (Roche Diagnostic, Indianapolis, IN, USA) was used. The procedure of hybridization was performed according to the manufacturer’s protocol. Digoxigenin-labeled cRNA probe was generated as described above. Digoxigenin-labeled sense cRNA probe was used as a negative control. The hybridization signals were visualized with chemiluminescence, which was recorded on X-ray films. The exposure time was 10 min.
Informed consent
The study protocol was approved by the Institutional Review Board of the hospital under the guidelines of the 1975 Declaration of Helsinki. Written informed consent was obtained from all patients.
DISCUSSION
Our previous bioinformatic analysis of GCRG123 showed that the predicted peptide sequence of GCRG123 has 80% homology with a human lamin-like protein (GenBank accession number, AAA36178)[9], therefore, we predicted that GCRG123 might be a gene that encoded a lamin-like protein. However, the results of the present study indicate that GCRG123 tends to be an active LINE-1 family member.
Retrotransposable elements, such as L1, Alu and endogenous retroviruses, make up some 45% of human DNA[16]. L1 is one of the most successful retrotransposons and occupies 17% of DNA[17]. In the early stages, because of the lack of any obvious correlation with cellular functions, retrotransposable elements were considered to be mere remnants of our genetic evolution and given the name ‘‘junk DNA’’[18]. However, recent data have revealed that some L1s do remain retrotransposition-competent. It has been shown that L1 is a functionally capable gene. For example, human embryonic stem cells can express endogenous L1 elements[19]. L1 can retrotranspose in neuronal precursors, and can consequently alter the expression of neuronal genes, which in turn, can influence neuronal cell fate in vitro[20]. L1 retrotransposition can also occur in non-dividing somatic cells[21].
A retrotransposition-competent, functional L1 element covers about 6.1 kb and contains a 5’ untranslated region, a 1-kb ORF1 that encodes a protein (p40) with RNA-binding ability, followed by a 3.8-kb ORF2 that codes for a protein (p150) with endonuclease and reverse transcriptase (RT) activities, and a cysteine-histidine-rich domain. The 3’ end of L1 is terminated by a short 3’ untranslated region and a poly(A) tail[22–26]. L1 likely integrates into the genomic DNA by a process called target primed reverse transcription and shapes mammalian genomes through many different ways, such as retrotransposition, transduction and gene expression alteration[27]. Although the overwhelming majority of L1s (about 99.8%) are inactive because of 5’ truncations, internal rearrangements and mutations, 80-100 retrotransposition-competent L1s have been predicted in humans. Remarkably, 84% of the retrotransposition capability of these elements has been shown to be present in six highly active L1s[15].
It has been proposed that retrotransposons modulate the expression of specific genes through a transcriptional interference-based mechanism. To exert such effect on neighboring genes, retrotransposons need not necessarily be full-length, nor retrotransposition-competent, but simply endowed with functional promoter elements[28]. Previous studies have shown that some non-full length L1 elements (even shorter than 500 bp) may yield L1 products such as L1 RNA or active RT[29–31]. L1 RNAs are also involved in extensive RNA splicing that can radically alter the diversity of expressed RNA forms from these elements, as well as influence their impact on gene expression upon genomic insertion[32].
The state-of-the-art understanding of L1 described above prompted us to propose that GCRG123 may be an active L1 family member, as supported by our findings. First, the GCRG123 sequence shared 92% homology with the conserved sequence of the highly active L1s (hot L1s). Second, GCRG123 can map to all chromosomes, which is similar to L1s. Third, GCRG123 can integrate in the intron or 5’ flanking region of some known genes, which suggests that it has the potential to adjust the function of these genes. Finally, in situ hybridization and Northern blot analysis at the mRNA level showed a distinct GCRG123 expression pattern in different kinds of gastric mucosa cells.
A lot of evidence has indicated that L1 is involved in a wide spectrum of diseases, including cancer[283334]. In colon cancer, the APC gene can be disrupted by somatic insertion of an L1 sequence into the last exon of the gene[35]. Takai et al have found that hypomethylation of L1 is detected in human hepatocellular carcinoma, but never in the surrounding liver tissues, whether or not liver cirrhosis is present[36]. Hypomethylation of L1 has also been revealed in urothelial carcinoma and prostate adenocarcinoma[37–39]. In malignant cells, there is a direct correlation between the hypomethylation of 5’ L1 sequences and the presence of L1 proteins, which suggests that elements with hypomethylated 5’ ends are transcriptionally active[40]. The hypomethylation of repetitive elements in cancer is directly linked to the neoplastic process and not a simple consequence of loss of growth control[41]. Previous studies have shown that L1 is essential for the proliferation of tumor cells. The functional knock-out of LINE-1 expression by RNAi reduces the proliferation and promotes the differentiation of tumorigenic human cells[2942]. Up till now, no data about the relationship between L1 and stomach cancer have been reported. The present study on the distinct expression pattern of GCRG123 in gastric signet-ring cell carcinoma may be the first to link an L1 family member to stomach oncogenesis.
During previous decades, several histological classification systems, such as those of Lauren, Nakamura and Ming, have been widely used for gastric adenocarcinoma. These systems agree that the intestinal/differentiated/expending type and diffuse/undifferentiated/infiltrative type differ greatly in terms of epidemiology, pathogenesis, genetic profile, and clinical outcome[843–46]. Signet-ring cell carcinoma belongs to the diffuse/undifferentiated/infiltrative type group. Nakamura has indicated that the morphological findings of undifferentiated type carcinoma under light and electron microscopy are similar to both the mucous cells of pyloric crypts and pyloric gland cells[44]. Interestingly, our study found that GCRG123 was over-expressed in pyloric gland cells, epithelia (surface mucous cells) and signet-ring cells, which correlated with Nakagawa’s description of undifferentiated carcinoma. The distinct GCRG123 expression patterns in intestinal-type adenocarcinoma and signet-ring cell carcinoma also support the concept that these two types of carcinoma have different genetic profiles. GCRG123 is over-expressed simultaneously in gastric epithelia, pyloric glands and signet-ring cell carcinoma, therefore, we believe that GCRG123 might be closely associated with gastric mucous secretion. Further studies are required to confirm this speculation.
COMMENTS
Background
Evidence indicates that a retrotransposition-competent, functional L1 element is involved in a wide spectrum of diseases, including cancer. A previous study has shown that a gene segment GCRG123 is down-regulated in human gastric intestinal-type adenocarcinoma. Intestinal- and diffuse-type gastric cancers differ in their epidemiology, pathogenesis, genetic profile and clinical outcome.
Research frontiers
To modulate the expression of specific genes, retrotransposons, such as L1, need not necessarily be full-length, nor retrotransposition-competent, but simply endowed with functional promoter elements. Some non-full length L1 elements could yield L1 products such as L1 RNA or active RT. L1 is essential for the proliferation of tumor cells. The functional knock-out of L1 expression reduces cell proliferation and promotes cell differentiation.
Innovations and breakthroughs
GCRG123 may be an active L1 family member, because of its sequence homology with the conserved sequence of the highly active L1s and its integration in the intron or 5’ flanking region of some known genes. In situ hybridization and Northern blot analysis showed that GCRG123 was up-regulated in signet-ring cell carcinoma. Besides, GCRG123 was also over-expressed in normal gastric epithelia and pyloric glands. The present study possibly represents the first to link an L1 family member to stomach oncogenesis.
Applications
Updated knowledge about L1 shifts the focus from protein-coding genes to repeated retroelements as mediators of abnormal biological processes. Functional L1 family members may be regarded as promising targets in the development of novel, differentiation-inducing approaches to cancer therapy. The distinct expression pattern of GCRG123 in gastric signet-ring cell carcinoma may provide a new area to further understand the genetic characteristics of gastric signet-ring cell carcinoma.
Terminology
LINE-1: highly repeated sequences, 6-8 K base pairs in length, which contain RNA polymerase II promoters. They also have an ORF that is related to the RT of retroviruses, but they do not contain long terminal repeats. Copies of the LINE 1 (L1) family form about 17% of the human genome.
Peer review
This is an interesting article. The title reflects the major contents of the article. The abstract gives a clear delineation of the research background, objectives, materials and methods, results and conclusions. The methods are innovative and systematic. A detailed description has been provided. The results provide sufficient experimental evidence or data from which scientific conclusions can be drawn. The discussion is well organized and an overall theoretical analysis is given. The most current literature is cited. Overall, a it is well-written and well-researched article.