Published online Jul 28, 2007. doi: 10.3748/wjg.v13.i28.3847
Revised: March 23, 2007
Accepted: March 26, 2007
Published online: July 28, 2007
AIM: To study immune response induced by foreign plasmid DNA after oral administration in mice.
METHODS: Mice were orally administered with 200 μg of plasmid pcDNA3 once and spleen was isolated 4 h and 18 h after administration. Total RNA was extracted from spleen and gene expression profile of BALB/c mice spleen was analyzed by using Affymetrix oligonucleotide GeneChip. Functional cluster analysis was conducted by GenMAPP software.
RESULTS: At 4 h and 18 h after oral plasmid pcDNA3 administration, a number of immune-related genes, including cytokine and cytokine receptors, chemokines and chemokine receptor, complement molecule, proteasome, histocompatibility molecule, lymphocyte antigen complex and apoptotic genes, were up-regulated. Moreover, MAPPFinder results also showed that numerous immune response processes were up-regulated. In contrast, the immunoglobulin genes were down-regulated.
CONCLUSION: Foreign plasmid DNA can modulate the genes expression related to immune system via the gastrointestinal tract, and further analysis of the related immune process may help understand the molecular mechanisms of immune response induced by foreign plasmid via the gastrointestinal tract.
- Citation: Liu JW, Cheng J. Molecular mechanism of immune response induced by foreign plasmid DNA after oral administration in mice. World J Gastroenterol 2007; 13(28): 3847-3854
- URL: https://www.wjgnet.com/1007-9327/full/v13/i28/3847.htm
- DOI: https://dx.doi.org/10.3748/wjg.v13.i28.3847
Foreign plasmid pcDNA3, which is a typical DNA vaccine vector, has an immune-stimulatory capacity ascribed to unmethylated CpG motif on its plasmid backbone[1,2]. Mammalian immune system recognizes synthetic oligodeoxyribonucleotides and bacterial DNA containing CpG dinucleotides in specific sequence contexts (CpG DNA)[3,4]. The immune responses to CpG DNA include stimulation of B cell proliferation, activation f macrophages, mono-cytes, and dendritic cells[3-6]. The activation of immune cells by CpG DNA results in the expression of several co-stimulatory molecules and secretion of a number of cytokines, including IL-12, IFN-γ, IL-6 and TNF-α[3-6].
The gastrointestinal tract (GIT) of mammals is the main portal of entry for foreign DNA and proteins[7,8]. We have documented the fate of orally administered plasmid DNA in the GIT of the mouse. Our previous work suggested that foreign plasmid DNA was not completely degraded in the GIT of mice[9]. Plasmid could be detected in almost all tissues 1 h after oral administration and the copies of plasmid in tissues changed with time. Foreign plasmid persisted transiently as fragments after feeding in the gut and organs[9]. However, foreign plasmid DNA could induce humoral and cell-mediated immune system in mice after administration via the GIT[10]. Plasmid DNA stimulated spleen lymphocyte proliferation and enforced phagocytic activity of macrophage in vivo[10]. It seemed the immunostimulatory activation mechanism is correlated to regulation of immune response gene in vivo.
The immune response was considered an ancient cellular defense mechanism against the activity of foreign genes. Apparently, the intestinal tract is not an absolute barrier against the uptake of macromolecules, such as plasmid DNA[11-14]. It will be interesting to investigate the mechanism of immune response and immune-related activated pathway induced by foreign plasmid DNA in vivo via the GIT. It will help further understanding metabolic mechanism and consequence of foreign DNA in vivo and may provide a critical clue for food safety and research on entry pathway of DNA vaccine into organism. Spleen has multiple hematologic and immunologic functions. It can not thoroughly elucidate regulation mechanism by single or several gene expression since immune system is a complicated regulation network. With the completion of the genome sequences of many model organisms and the advent of DNA microarray technology, simultaneous monitoring of the transcriptional levels of thousands of genes in a genome has become possible. In this study, we employed Affymetrix array technology to characterize the gene expression profile in the spleen after oral administration of foreign plasmid pcDNA3 in mice. It can provide new sight into the overall cellular and molecular consequence of foreign DNA absorbed via the gastrointestinal tract in the mammal systems.
Plasmid was propagated in E. coli DH5α, purified using EndoFree Plasmid Maxi or Mega kit (Qiagen, Hilden, Germany), diluted in TE buffer and passed over a detoxi-gel column (Piere, Rockford, IL, USA). The content of endotoxin was estimated by using limulus amebocyte lysate test (QCL-1000 test, BioWhittaker East Rutherford, NJ, USA). Before administration, 2 mg of the plasmid pcDNA3 was diluted in 2 mL of pyrogen-free NaCl. The content of endotoxin after final dilution never exceeded 0.25 EU/mL.
Six-week-old male Balb/c mice were obtained from Shanghai Experimental Animal Center (Shanghai, China) and maintained in a specific pathogen-free condition. Mice were orally administered by pipette with 200 μg of plasmid pcDNA3 in 200 μL of saline once and sacrificed at 4 h and 18 h later. The spleen was isolated and cleaned by RNase-free saline. Three spleens of the same group mice were mixed together and stored in RNA at 4°C overnight and maintained at -20°C.
Total RNA was extracted from the spleen using Trizol (Invitrogen, Carlsbad, CA). The quality of RNA samples was determined by agarose gel electrophoresis and staining with ethidium bromide; the 18S and 28S RNA bands were visualized under UV light. To perform microarray hybridization, two independent extractions of RNA were obtained from the plasmid pcDNA3 groups and the control group.
Affymetrix MOE4 GeneChips, encompassing about 22 690 genes and ESTs on one array were processed according to the manufacturer’s recommendations. Equal amount of the RNA from each group was taken for purification of poly (A) mRNA by using a poly (A) purification kit (Promega, Madison, WI). RNA samples were reverse-transcribed with poly dT oligonucleotide attached to a sequence of the T7 promoter region, digested with RNase H, and copied into dsDNAs (SuperScript Choice System, Invitrogen). In vitro RNA transcription was performed to incorporate biotin-labeled ribonucleotides into the cRNA transcripts using an RNA transcript labeling kit (Enzo Biochem, Farmingdale, NY). Labeled cRNAs were purified and analyzed by agarose gel electrophoresis to confirm a size distribution ranging from 500 to 1200 bases. These cRNAs were fragmented to sizes ranging from 50 to 200 bases by heating at 94°C for 35 min, and then were used for separate hybridization to a rat Genome U34A Array (Affymetrix, Santa Clara, CA) according to the manufacturer’s instructions, with a prior quality assay using Test2 Array probe chips. After hybridization and subsequent washing using the Affymetrix Fluidics station 400, the bound RNAs were stained with streptavidin phycoerythrin, and the signals were amplified with a fluorescent-tagged antibody to streptavidin. Fluorescence was measured using the Affymetrix scanner, and the results were analyzed using the GeneChip Analysis Suite software. Log2 of the respective normalization factor was added to log2 of the ratio for each spot within the array, in such a way that the average log-transformed ratio was equal to zero.
Before applying the cut-off filtering criterion, the gene expression values selected as described above were subjected to functional cluster analysis using GenMAPP and MAPPFinder[15,16], a software that creates a global gene expression profile from microarray data by integrating the annotations of the Gene Ontology (GO) Project (Http://http://www.geneontology.org) with the free software package GenMAPP (Http://http://www.genmapp.org). Using the GenMAPP Expression Dataset Manager tool, we converted the median of gene expression values (.xls) into an expression data set file (.gex) and defined the criteria for meaningful gene changes in expression. The criteria were set up to consider fold ≥ 2 (|SLR| ≥ 1, SLR is equal to log2fold) which meant significantly up-regulated and down-regulated genes. Having created the expression Dataset file, we obtained functional cluster results using the MAPPFinder program. MAPPFinder builds a local copy of the GO hierarchy using the three Ontology files (process, function and component) available from GO. The links between GO terms and genes in the expression data set were created through Affymetrix probe set. For each term, MAPPFinder calculates (1) a first set of percentages for the genes specifically associated with that term of the GO hierarchy, (2) a second set of percentages for the total number of genes associated with that term and all its “children”, and (3) a z score (i.e. the level of confidence that a term has more or less genes meeting the criterion than those would be expected by chance).
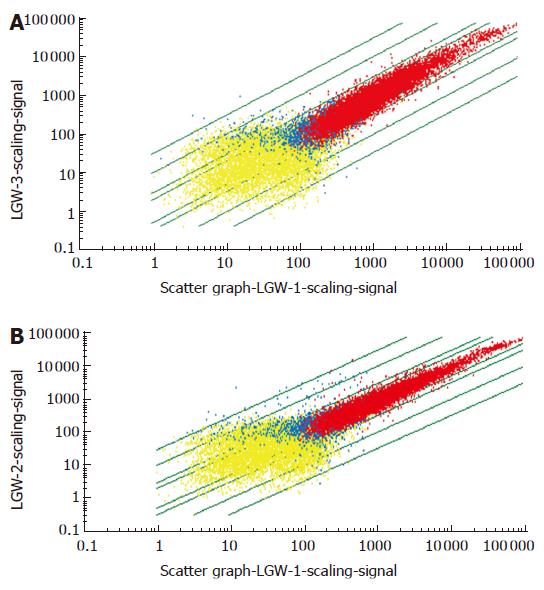
To investigate a systemic response to plasmid pcDNA3, Balb/c mice were orally administered with plasmid pcDNA3, and spleen RNA was harvested at 4 h and 18 h after oral administration. These RNA samples were hybridized to Affymetrix MOE4 oligonucleotide arrays containing 22 690 oligonucleotide probe sets, each representing an expressed genes. Therefore, the array represented roughly two-third of the protein coding capacity of mouse genome. The general information of gene expression is presented in Figure 1A and B. A comparison of spleen gene between the plasmid pcDNA3 group and the control group revealed that foreign plasmid DNA could cause global change in gene expression in the spleen. Compared to the control group, 1803 genes were up-regulated and 578 genes were down-regulated at 4 h after oral administration. While 1620 genes were up-regulated and 143 genes were down-regulated at 18 h after oral administration. The result of total differential gene expression is presented in Table 1.
| Categories | Number of up-regulatedgenes | Number of down-regulatedgenes | ||
| 4 h | 18 h | 4 h | 18 h | |
| |SLR| < 1 | 1527 | 1225 | 131 | 73 |
| |SLR| ≥ 1 | 276 | 395 | 447 | 70 |
| |SLR| ≥ 2 | 64 | 188 | 92 | 17 |
| |SLR| ≥ 3 | 21 | 95 | 60 | 10 |
| |SLR| ≥ 4 | 4 | 32 | 27 | 7 |
| Total | 1803 | 1620 | 578 | 143 |
We used MappFinder to assign the differentially expressed genes which changed greater than 2-fold (|SLR| ≥ 1) to non-mutually exclusive categories regarding biological process, molecular function, and/or cellular components. MappFinder calculates (1) the percentages of genes meeting the user-defined criterion, and (2) the z score for each GO term and local term. MAPPFinder was able to connect 211 of the 292 significantly up-regulated genes (SLR > 1) and 296 of 414 significantly down-regulated genes to GO terms. Interestingly, the processes which significantly up-regulated the genes involved at 4 h included immune response, defense response, innate immune response, inflammatory response, apoptotic, and pyrimidine nucleotide metabolism. At 18 h, the up-regulated process mainly included digestion, heat stress response, RNA process and metabolism, and protein metabolism. While in down-regulated genes, the process mainly included humoral response, cell growth and regulation, protein modification, phosphoric acid metabolism. The processes which significantly down-regulated genes involved at 18 h mainly included humoral response. At 4 h, up-regulated genes mainly took place in ribosome, ribonucleoprotein complex, mitochondrial inner membrane and cytosol, while at 18 h the up-regulated genes mainly took place on cell membrane. The down-regulated genes took place in the nucleus at 4 h and in immunoglobulin complex at 18 h.
The results (Table 2) showed that a number of immune genes were up-regulated. These genes mainly included cytokine and cytokine receptors, chemokine and chemo-kine receptors, complement molecule, proteasome, histocompatibility molecule, lymphocyte antigen complex and apoptotic genes. The down-regulated genes were mainly immunoglobulin genes.
| Probe set | Gene symbol | 4 h (SLR) | 18 h(SLR) | Gene title |
| 1423754_at | 1110004C05Rik | 0.9 | 0.8 | RIKEN cDNA 1110004C05 gene |
| 1425156_at | 9830147J24Rik | 0.4 | 1.6 | RIKEN cDNA 9830147J24 gene |
| 1452428_a_at | B2m | 0.5 | 0.4 | beta-2 microglobulin |
| 1418021_at | C4 | 0.3 | 0.3 | Complement component 4 (within H-2S) |
| 1418126_at | Ccl5 | 0.3 | 0.3 | Chemokine (C-C motif) ligand 5 |
| 1419609_at | Ccr1 | 1.1 | 0.4 | Chemokine (C-C motif) receptor 1 |
| 1418930_at | Cxcl10 | 0.3 | 1.2 | Chemokine (C-X-C motif) ligand 10 |
| 1417876_at | Fcgr1 | 1.5 | 0.4 | Fc receptor, IgG, high affinity I |
| 1431591_s_at | G1p2 | 1.7 | 1.6 | interferon, alpha-inducible protein |
| 1420549_at | Gbp1 | 0.6 | 0.7 | Guanylate nucleotide binding protein 1 |
| 1435906_x_at | Gbp2 | 0.7 | 0.2 | Guanylate nucleotide binding protein 2 |
| 1421596_s_at | H28 | 1.9 | 1.4 | Histocompatibility 28 |
| 1425917_at | H28 | 1.5 | 1.8 | Histocompatibility 28 |
| 1451721_a_at | H2-Ab1 | 0.3 | 0.2 | Histocompatibility 2, class II antigen A, beta 1 |
| 1426324_at | H2-D1 | 0.7 | 0.7 | Histocompatibility 2, D region locus 1 |
| 1424948_x_at | H2-K | 0.3 | 0.6 | Histocompatibility 2, K region |
| 1425336_x_at | H2-K | 0.3 | 0.3 | Histocompatibility 2, K region |
| 1452544_x_at | H2-K | 0.5 | 0.3 | Histocompatibility 2, K region |
| 1421358_at | H2-M3 | 0.5 | 0.2 | Histocompatibility 2, M region locus 3 |
| 1449875_s_at | H2-T10 | 0.6 | 0.4 | histocompatibility 2, T region locus 10 |
| 1449556_at | H2-T23 | 0.3 | 0.2 | Histocompatibility 2, T region locus 23 |
| 1419603_at | Ifi16 | 4.6 | 4.2 | Interferon, gamma-inducible protein 16 |
| 1421551_s_at | Ifi202b | 1.5 | 0.9 | Interferon activated gene 202B |
| 1452348_s_at | Ifi205 | 0.5 | 0.4 | Interferon activated gene 205 |
| 1452349_x_at | Ifi205 | 0.8 | 0.7 | Interferon activated gene 205 |
| 1450783_at | Ifit1 | 0.8 | 0.6 | Interferon-induced protein with tetratricopeptide repeats 1 |
| 1449025_at | Ifit3 | 1.1 | 0.8 | Interferon-induced protein with tetratricopeptide repeats 3 |
| 1418219_at | Il15 | 1.6 | 1.4 | Interleukin 15 |
| 1416296_at | Il2rg | 0.5 | 0.6 | Iinterleukin 2 receptor, gamma chain |
| 1449399_a_at | Il1b | 0.9 | 0.4 | Interleukin 1 beta |
| 1421322_a_at | Isgf3g | 0.3 | 0.4 | Iinterferon-dependent positive acting transcription factor 3 gamma |
| 1425436_x_at | Klra3 | 0.7 | 0.1 | Killer cell lectin-like receptor, subfamily A, member 3 |
| 1417185_at | Ly6a | 0.9 | 0.9 | Lymphocyte antigen 6 complex, locus A |
| 1421571_a_at | Ly6c | 0.7 | 0.3 | Lymphocyte antigen 6 complex, locus C |
| 1416930_at | Ly6d | 0.6 | 0.5 | Lymphocyte antigen 6 complex, locus D |
| 1422089_at | Ncr1 | 1.4 | 1.0 | Natural cytotoxicity triggering receptor 1 |
| 1453196_a_at | Oasl2 | 2.1 | 1.9 | 2'-5' oligoadenylate synthetase-like 2 |
| 1422962_a_at | Psmb8 | 0.4 | 0.3 | Proteosome (prosome, macropain) subunit, beta type 8 (large multifunctional protease 7) |
| 1450696_at | Psmb9 | 0.8 | 0.6 | Proteosome (prosome, macropain) subunit, beta type 9 (large multifunctional protease 2) |
| 1417056_at | Psme1 | 0.5 | 0.4 | Proteasome (prosome, macropain) 28 subunit, alpha |
| 1418131_at | Samhd1 | 0.6 | 0.2 | SAM domain and HD domain, 1 |
| 1421812_at | Tapbp | 0.3 | 0.5 | TAP binding protein |
| 1425324_x_at | Igh-4 | -1.7 | -2.1 | Immunoglobulin heavy chain 4 (serum IgG1) |
| 1429381_x_at | Igh-VJ558 | -1.2 | -1.4 | Immunoglobulin heavy chain (J558 family) |
| 1451632_a_at | Igh-1 | -2 | -2.5 | Immunoglobulin heavy chain 1 (serum IgG2a) |
Our data provided a global view in differential genes expression after administration of foreign plasmid pcDNA3 via the gastrointestinal tract. The result of function cluster analysis of differential genes expression by MappFinder clearly showed that oral administration of plasmid pcDNA3 induced a number of immune response genes, mainly including cytokine and cytokine receptors, chemokine and chemokines receptors, complement molecule, proteasome, histocompatibility molecule, lymphocyte antigen complex and apoptotic genes. The down-regulated genes were mainly immunoglobulin genes. Some groups of key genes which participate in immune response are discussed below.
Nfkbia and Nfkbib genes were up-regulated at 4 h and Nfkb2 and Nfkbia genes were up-regulated at 18 h after oral administration of plasmid pcDNA3. Recent studies on CpG-mediated immune activation suggest that CpG functions through Toll-like receptor 9 (TLR9) to deliver signals intracellularly, culminating in activation of transcription factors NF-κB and AP-1, which, in turn, enhances a number of genes previously implicated in control of various immune cells[17-19]. The expression of both Nfkbia (IkBa/mad3) and Nfkbib (IkBb) increased after plasmid administration. The function of NF-κB can therefore be linked to that of a second messenger molecule through its ability to transduce upstream signals from the cytoplasm into the nucleus in activated cells in TLR9 pathway. The IKK complex which is activated by CpG DNA phosphorylates IκB, leading to proteasome-mediated IκB degradation, and then NF-κB translocates from the cytosol to the nucleus to mediate the expression of pro-inflammatory cytokines[20]. MyD88 was also up-regulated after plasmid pcDNA3 administration. Signaling by CpG DNA through TLR9 requires participation of the adaptor protein MyD88 and results in activation of common transcription factors NF-κB and AP-1. We can deduce that plasmid pcDNA3 may activate innate immune response through TLR9-NF-κB pathway.
The results presented in this study clearly showed that plasmid pcDNA3 induced a number of cytokine and cytokine receptor genes expression. Compared with humans, mice may be classified as ‘high’ CpG-DNA responders[21]. In this result, systemic challenge of mice with plasmid pcDNA3 causes a transient ‘cytokine storm’ in the spleen; substantial concentrations of pro-inflammatory cytokines, including chemokines and cytokines, such as chemokine (C-C motif) ligand 12 (Ccl12), interferon, gamma-inducible protein 16 (Ifi16), chemokine (C-C motif) ligand 3 (Ccl3). At 4 h, Ccl12 appeared as the most strongly induced genes, showing SLR of 5.6. Chemokines play a key role in the balance of Th1/Th2 response. Chemokines, such as MCP-1 (Ccl12), RANTES (Ccl5) and IP10 (Ccl10), are related to Th1 response. These chemokines can promote Th1 cell immune response and inhibit Th2 activity by decreasing IL-4 expression. Previous studies have demonstrated that CpG ODN induces a significant increase in chemokine mRNA levels at the site of injection and draining lymph nodes within 6 h of in vivo administration[22], thereby indicating that plasmid pcDNA3 can promote pro-Th1 response by inducing some chemokines expression. We also found a number of cytokine genes, such as IFN-γ, IL-2, interferon gamma-inducible protein, interferon-induced protein, Il2rg and Il2rb, were induced after oral administration. Il2r is a key receptor during T cell activation. The binding between IL-2 and IL2r improves the production of IL-2 that subsequently induces T cell activation and proliferation. Overall, plasmid pcDNA3 induces a Th1-like pattern of cytokine production dominated by IFN-γ, IL-2 and pro-Th1 chemokines.
The results showed that plasmid pcDNA3 up-regulated a series of hiscompatibility genes, including H2-Ab1, H2-D1, H2-K, H2-M3, H2-T10, H2-T23 and H28. Hiscompatibility antigen is encoded by H2 and H2 complex in mice. These genes encode MHC I molecule, which is the principal antigen in the immune response. MHC class I molecules are loaded with proteins generated in the cytosol. It is deduced that the innate immune cells-macrophages and DCs-are acutely activated by plasmid pcDNA3, resulting in the up-regulation of MHC class I and enhancing presentation capacity of antigen T cell via MHC class I pathway[21].
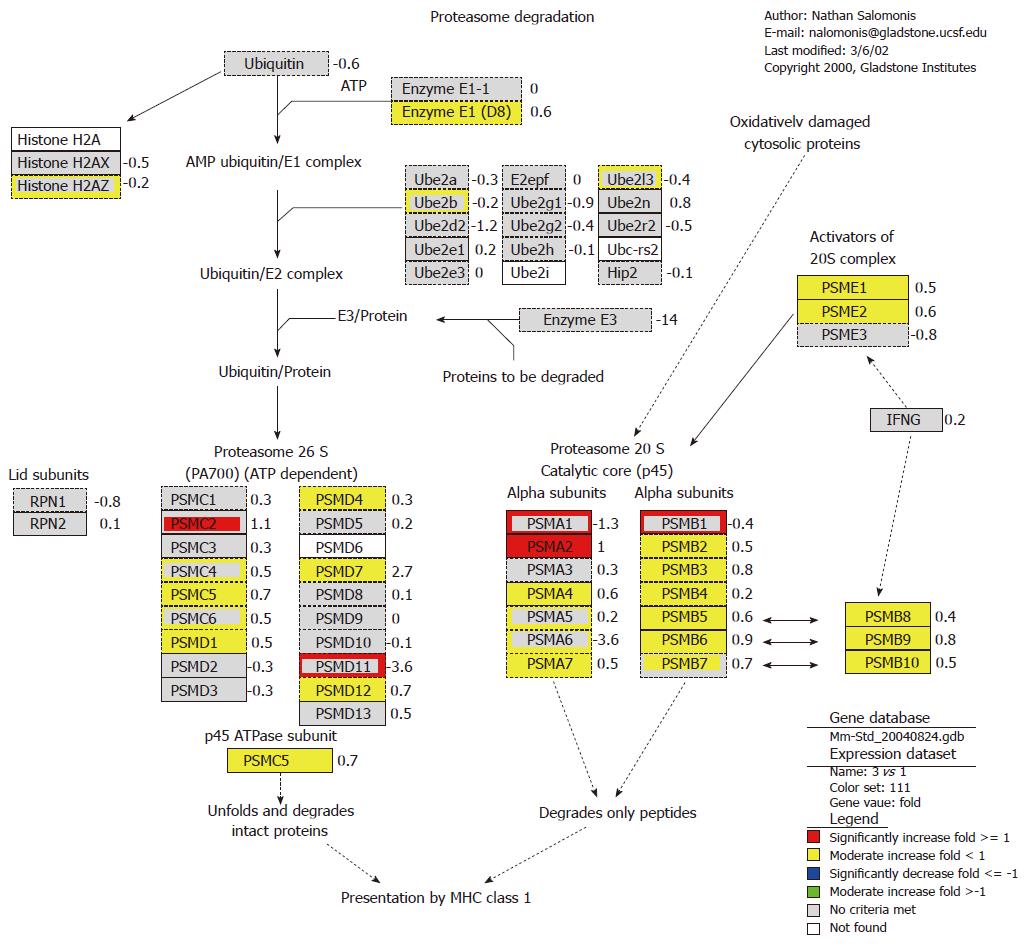
The result also showed that foreign plasmid pcDNA3 up-regulated the expression of proteasome gene, such as Psmb8, Psmb9 and Psme1 (Figure 2). Almost all genes in α and β subunit of 20S proteasome were up-regulated at 4 h, and part of genes in 26S proteasome were also up-regulated. The proteasome plays a straightforward but critical role in the function of the adaptive immune system. The peptide antigens displayed by the major histocompatibility complex class I (MHC) proteins on the surface of antigen-presenting cells are products of proteasomal degradation of proteins originated by the invading pathogen. The genes of proteasome 20S subunits were almost induced, while ubiquitin was significantly suppressed. This indicated that plasmid pcDNA3 could induce immune response in endogenous antigen pathway. It can be concluded that foreign plasmid pcDNA3 may activate major hiscompatibility antigen and 20S proteasome genes in ubiquitin-proteasome pathway, and so improve antigen presentation ability to induce T cell activation.
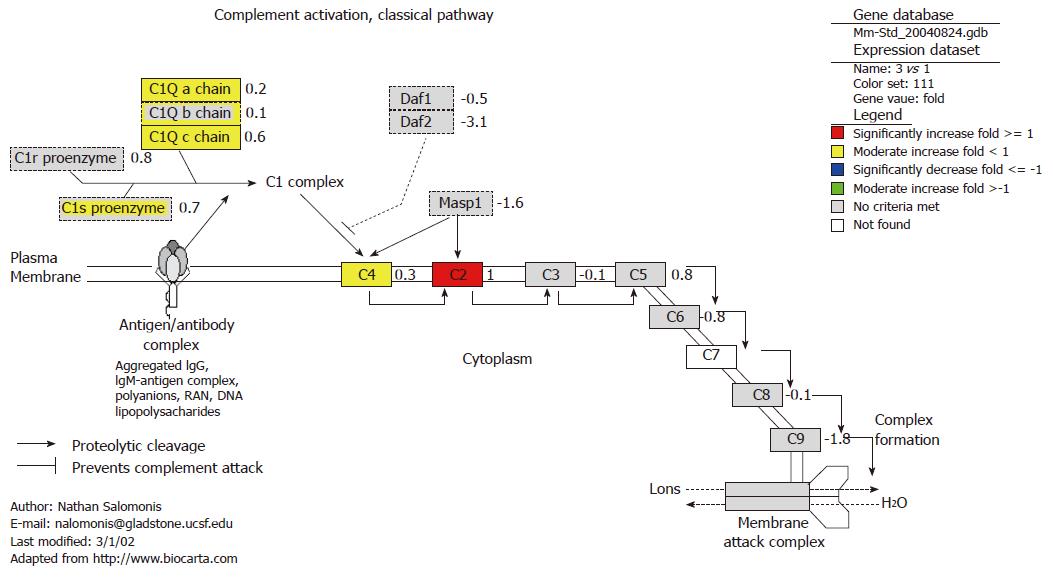
All genes, such as C2, C4, C1s proenzyme and C1q which involve in prophase of complement classical pathway, were activated at 4 h after oral administration of plasmid pcDNA3 (Figure 3). Complement component 4 (C4) was up-regulated at both 4 h and 18 h. The classical pathway of the complement system is a major effector of the humoral branch of the human immune response. The complement system has many other important roles in mediating and enhancing the immune response against a wide variety of invaders, such as foreign plasmid pcDNA3. The activation of complement classical pathway demonstrated the activation of this pathway occurred in the early phase before the formation of C3 convertase. Since activation of the complement system is a key part of the immune system, the complement system is a part of the defense system against invading foreign plasmid DNA, which stimulates phagocytosis of plasmid pcDNA3 and induces an inflammatory response.
Our results also showed that foreign plasmid pcDNA3 could induce expression of a number of apoptotic genes, including Casp3, Birc2, Birc3, Cycs, Bid, Bax, Nfkbia, Tnfrsf1b and Traf2. On contrary, the Birc4 genes were inhibited at 4 h. The mechanism of CpG DNA-mediated apoptosis is still not clear, but it has been reported that CpG DNA can inhibit apoptosis by caspase-dependent and -independent pathways. Wang et al[23] investigated the role of unmethylated mitogenic CpG motifs in regulating Fas-mediated apoptosis in primary murine B cells[23]. Unmethylated CpG motif protected CD40L-stimulated B cells from Th1-CMC and apoptosis mediated by Fas-specific antibody. It suggest that Fas-mediated apoptosis requires minimum up-regulation of surface Fas expression and that CpG motifs protect B cells from Fas-mediated apoptosis by decreasing surface Fas expression.
We found that expression of IAP1 and IAP2 genes was up-regulated. It has been reported that activation of NF-κB can induce expression of cIAP1 and cIAP2[23], which is similar to our result. The result that plasmid pcDNA3 up-regulated Cycs , Bid and Bax genes expression suggested that plasmid pcDNA3 regulated cell apoptosis via a mitochondrial mechanism. Caspase 3, which is a key apoptosis factor, was found be induced. Caspase 3 is responsible for the cleavage of the key cellular proteins that lead to the typical morphological changes observed in cells undergoing apoptosis. Moreover, tumor necrosis factor receptor gene (Tnfrsf1b) was found to be up-regulated. Tnfrs1b, a member of TNF receptor superfamily, can complex with TNF receptor 1, and then regulate anti-apoptosis proteins c-IAP1 and c-IAP2. The role of c-IAP1 is to reinforce the apoptotic pathway mediated by TNF. CpG DNA has been shown to activate NF-κB and to induce the transcription of Bcl-XL. These factors may synergize with DNA repair enzyme, DNA-PK, to inhibit cell death at mucosal sites mediated by the administration of CpG DNA[24]. Plasmid pcDNA3 can inhibit cell apoptosis, contributing to the generation of a sustained immune response.
In a previous report, we found that oral administration of foreign plasmid DNA can induce humoral and cell-mediated immunity in mice[10]. In this report, we demonstrated, probably for the first time, by microarray that orally administered plasmid pcDNA3 can induce global immune-related genes in vivo. The results showed that a large number of immune genes were up-regulated, while a few immune genes were down-regulated. MAPPFinder results also revealed that plasmid pcDNA3 induced a large number of immune-related processes. It suggested that oral administration of plasmid pcDNA3, which is considered a foreign immune stimulator, sharply up-regulated immune gene in the spleen. Interestingly, we found that the immune genes induced at 4 h were more than those at 18 h, indicating that foreign plasmid-induced immune genes expressions mainly take place at the early stage and the induction become weaker gradually.
It can be considered that foreign plasmid pcDNA3 acts as “danger signal” recognized by host innate immune system via the gastrointestinal tract. The immuno-stimulatory plasmid pcDNA3 has been ascribed to unmethylated CpG motif on its skeletal. Because CpG motifs are abundant in bacterial and viral genomes, they have been suggested as a common recognition structure, triggering cells of the vertebrate immune system[25-30]. Foreign plasmid DNA could be incompletely degraded in the gastrointestinal tract. It has been reported that foreign DNA (M13, plasmid) can be absorbed by the gastrointestinal tract in a rapid process[11]. Foreign DNA fragment remaining in the intestinal tract could potentially be absorbed through the intestinal digesta either directly by epithelial cells or by antigen-presenting cells of the immune system. Part of orally administered plasmid possibly passes through the mucosal barrier and enters systemically through the blood stream to the liver and spleen, where immune responses are stimulated. Foreign DNA in the peripheral blood and spleen was predominantly present in leukocytes, perhaps because of their defense functions. The leukocytes carrying the foreign DNA had possibly migrated from the Peyer’s patches in the gut wall to the bloodstream and spleen[11]. It is suggested that most of foreign DNA would be phagocytised by tissue macrophages, dendritic cells or other terminally differentiated phagocytes of the immune system. Regardless of the mechanism, orally administered foreign plasmid pDNA53 can up-regulate substantially immune gene expression in the spleen. These results provide us with a systematic and comprehensive insight into the mechanism of uptake and consequences of foreign plasmid pDNA3 administered via the gastrointestinal tract and also provide key clues for gene therapy and DNA vaccine.
The gastrointestinal tract is the main portal of entry of foreign DNA into organisms. Foreign plasmid can be absorbed by gastrointestinal tract and distribute in different tissues quickly, surviving in fragment form. The effect of foreign DNA ingested via the gastrointestinal tract on the organism system has hardly been investigated. Aside from its function of encoding the genetic material, DNA can have direct immune-stimulatory effects. It will be interesting to investigate the mechanism of immune response and immune-related activated pathway induced by foreign plasmid DNA in vivo via the gastrointestinal tract.
In previous studies, the DNA of bacteriophage M13 and plasmid vector have been shown to persist in fragmented form in minute amounts in different parts of the gastrointestinal tract and to gain access to cells of the intestinal wall, to the Peyer’s patches, to peripheral white blood cells, and to cells in the spleen and liver. Furthermore, food-ingested DNA can transgress the placental barrier in pregnant mice, but only a few cells take up the foreign DNA. Apparently, the gastrointestinal tract is not an absolute barrier against the uptake of macromolecules, such as foreign DNA.
With the advent of DNA microarray technology, simultaneous monitoring of the transcriptional levels of thousands of genes in a genome has become possible. We employed Affymetrix array technology to characterize immune gene expression profile in the spleen after oral administration of foreign plasmid pcDNA3 in mice. It provides new insight into the overall cellular and molecular consequence of foreign DNA absorbed via the gastrointestinal tract in the mammal systems.
Foreign plasmid pDNA3 given orally can up-regulate substantial immune genes expression in the spleen. These results provide us with a systematic and comprehensive insight into the mechanism of uptake and consequences of foreign plasmid pDNA3 administered via the gastrointestinal tract and also provide key clues for gene therapy and DNA vaccine.
This is an interesting manuscript showing that oral administration of foreign DNA alters gene expression in mice. An up-regulation of more than 100 immune-related genes and a down-regulation of immunoglobulin genes were observed.
S- Editor Liu Y L- Editor Kumar M E- Editor Ma WH
| 1. | Johansson E, Wallgren P, Fuxler L, Domeika K, Lefèvre F, Fossum C. The DNA vaccine vector pcDNA3 induces IFN-alpha production in pigs. Vet Immunol Immunopathol. 2002;87:29-40. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 12] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 2. | Magnusson M, Johansson E, Berg M, Eloranta ML, Fuxler L, Fossum C. The plasmid pcDNA3 differentially induces production of interferon-alpha and interleukin-6 in cultures of porcine leukocytes. Vet Immunol Immunopathol. 2001;78:45-56. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 25] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 3. | Krieg AM. CpG motifs in bacterial DNA and their immune effects. Annu Rev Immunol. 2002;20:709-760. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1936] [Cited by in RCA: 1945] [Article Influence: 84.6] [Reference Citation Analysis (0)] |
| 4. | Klinman DM. Immunotherapeutic uses of CpG oligodeoxynucleotides. Nat Rev Immunol. 2004;4:249-258. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 725] [Cited by in RCA: 776] [Article Influence: 37.0] [Reference Citation Analysis (0)] |
| 5. | Klinman DM. Use of CpG oligodeoxynucleotides as immunoprotective agents. Expert Opin Biol Ther. 2004;4:937-946. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 66] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 6. | Ishii KJ, Gursel I, Gursel M, Klinman DM. Immunotherapeutic utility of stimulatory and suppressive oligodeoxynucleotides. Curr Opin Mol Ther. 2004;6:166-174. [PubMed] |
| 7. | Doerfler W, Schubbert R, Heller H, Hertz J, Remus R, Schröer J, Kämmer C, Hilger-Eversheim K, Gerhardt U, Schmitz B. Foreign DNA in mammalian systems. APMIS Suppl. 1998;84:62-68. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 13] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 8. | Doerfler W, Remus R, Müller K, Heller H, Hohlweg U, Schubbert R. The fate of foreign DNA in mammalian cells and organisms. Dev Biol (Basel). 2001;106:89-97; discussion 143-160. [PubMed] |
| 9. | Liu JW, Shi YH, Le GW. Metabolic kinetics of foreign plasmid DNA uptake via gastrointestinal tract in mice. Shijie Huaren Xiaohua Zazhi. 2004;12:1108-1113. |
| 10. | Liu JW, Shi YH, Le GW. Effect of oral administration of foreign plasmid DNA on immune fuction in mice. Shijie Huaren Xiaohua Zazhi. 2004;12:2614-2617. |
| 11. | Schubbert R, Renz D, Schmitz B, Doerfler W. Foreign (M13) DNA ingested by mice reaches peripheral leukocytes, spleen, and liver via the intestinal wall mucosa and can be covalently linked to mouse DNA. Proc Natl Acad Sci USA. 1997;94:961-966. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 206] [Cited by in RCA: 181] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 12. | Doerfler W, Hohlweg U, Müller K, Remus R, Heller H, Hertz J. Foreign DNA integration--perturbations of the genome--oncogenesis. Ann N Y Acad Sci. 2001;945:276-288. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 35] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 13. | Müller K, Heller H, Doerfler W. Foreign DNA integration. Genome-wide perturbations of methylation and transcription in the recipient genomes. J Biol Chem. 2001;276:14271-14278. [PubMed] |
| 14. | Remus R, Kämmer C, Heller H, Schmitz B, Schell G, Doerfler W. Insertion of foreign DNA into an established mammalian genome can alter the methylation of cellular DNA sequences. J Virol. 1999;73:1010-1022. [PubMed] |
| 15. | Dahlquist KD, Salomonis N, Vranizan K, Lawlor SC, Conklin BR. GenMAPP, a new tool for viewing and analyzing microarray data on biological pathways. Nat Genet. 2002;31:19-20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 700] [Cited by in RCA: 669] [Article Influence: 29.1] [Reference Citation Analysis (0)] |
| 16. | Doniger SW, Salomonis N, Dahlquist KD, Vranizan K, Lawlor SC, Conklin BR. MAPPFinder: using Gene Ontology and GenMAPP to create a global gene-expression profile from microarray data. Genome Biol. 2003;4:R7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 613] [Cited by in RCA: 639] [Article Influence: 29.0] [Reference Citation Analysis (0)] |
| 17. | Zhao H, Hemmi H, Akira S, Cheng SH, Scheule RK, Yew NS. Contribution of Toll-like receptor 9 signaling to the acute inflammatory response to nonviral vectors. Mol Ther. 2004;9:241-248. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 62] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 18. | Verthelyi D, Zeuner RA. Differential signaling by CpG DNA in DCs and B cells: not just TLR9. Trends Immunol. 2003;24:519-522. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 77] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 19. | Takeshita F, Gursel I, Ishii KJ, Suzuki K, Gursel M, Klinman DM. Signal transduction pathways mediated by the interaction of CpG DNA with Toll-like receptor 9. Semin Immunol. 2004;16:17-22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 140] [Cited by in RCA: 141] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 20. | Akira S, Hemmi H. Recognition of pathogen-associated molecular patterns by TLR family. Immunol Lett. 2003;85:85-95. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 803] [Cited by in RCA: 825] [Article Influence: 37.5] [Reference Citation Analysis (0)] |
| 21. | Wagner H. Interactions between bacterial CpG-DNA and TLR9 bridge innate and adaptive immunity. Curr Opin Microbiol. 2002;5:62-69. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 142] [Cited by in RCA: 143] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 22. | Takeshita S, Takeshita F, Haddad DE, Ishii KJ, Klinman DM. CpG oligodeoxynucleotides induce murine macrophages to up-regulate chemokine mRNA expression. Cell Immunol. 2000;206:101-106. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 59] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 23. | Wang Z, Karras JG, Colarusso TP, Foote LC, Rothstein TL. Unmethylated CpG motifs protect murine B lymphocytes against Fas-mediated apoptosis. Cell Immunol. 1997;180:162-167. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 27] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 24. | Yi AK, Hornbeck P, Lafrenz DE, Krieg AM. CpG DNA rescue of murine B lymphoma cells from anti-IgM-induced growth arrest and programmed cell death is associated with increased expression of c-myc and bcl-xL. J Immunol. 1996;157:4918-4925. [PubMed] |
| 25. | Krieg AM. Immune effects and mechanisms of action of CpG motifs. Vaccine. 2000;19:618-622. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 121] [Cited by in RCA: 119] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 26. | Krieg AM, Yi AK, Matson S, Waldschmidt TJ, Bishop GA, Teasdale R, Koretzky GA, Klinman DM. CpG motifs in bacterial DNA trigger direct B-cell activation. Nature. 1995;374:546-549. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2735] [Cited by in RCA: 2617] [Article Influence: 87.2] [Reference Citation Analysis (0)] |
| 27. | Chace JH, Hooker NA, Mildenstein KL, Krieg AM, Cowdery JS. Bacterial DNA-induced NK cell IFN-gamma production is dependent on macrophage secretion of IL-12. Clin Immunol Immunopathol. 1997;84:185-193. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 226] [Cited by in RCA: 218] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 28. | Stacey KJ, Sweet MJ, Hume DA. Macrophages ingest and are activated by bacterial DNA. J Immunol. 1996;157:2116-2122. [PubMed] |
| 29. | Jakob T, Walker PS, Krieg AM, Udey MC, Vogel JC. Activation of cutaneous dendritic cells by CpG-containing oligodeoxynucleotides: a role for dendritic cells in the augmentation of Th1 responses by immunostimulatory DNA. J Immunol. 1998;161:3042-3049. [PubMed] |
| 30. | Doerfler W, Schubbert R. Uptake of foreign DNA from the environment: the gastrointestinal tract and the placenta as portals of entry. Wien Klin Wochenschr. 1998;110:40-44. [PubMed] |