Published online Apr 7, 2007. doi: 10.3748/wjg.v13.i13.1975
Revised: January 17, 2007
Accepted: March 19, 2007
Published online: April 7, 2007
AIM: To determine the genotype distribution of hepatitis B virus (HBV) with a newly oligonucleotide chip assay among the HBV carriers in Eastern China.
METHODS: An assay using oligonucleotide chip was developed for detection of HBV genotypes in serum samples from HBV DNA-positive patients in Eastern China. This method is based on the principle of reverse hybridization with Cy5-labeled amplicons hybridizing to type-specific oligonucleotide probes that are immobilized on slides. The results of 80 randomly chosen sera were confirmed by direct sequencing.
RESULTS: HBV genotype B, C and mixed genotype were detected in 400 serum samples, accounting for 8.3% (n = 33), 83.2% (n = 333), and 8.5% (n = 34), respectively. The evaluation of the oligonucleotide assay showed 100% concordance with the amplicon phylogenetic analysis except 9 mixed genotype infections undetected by sequencing.
CONCLUSION: The study indicates that HBV genotype C and B prevail in the Eastern China. It is suggested that the oligonucleotide chip is a reliable and convenient tool for the detection of HBV genotyping.
- Citation: Tang XR, Zhang JS, Zhao H, Gong YH, Wang YZ, Zhao JL. Detection of hepatitis B virus genotypes using oligonucleotide chip among hepatitis B virus carriers in Eastern China. World J Gastroenterol 2007; 13(13): 1975-1979
- URL: https://www.wjgnet.com/1007-9327/full/v13/i13/1975.htm
- DOI: https://dx.doi.org/10.3748/wjg.v13.i13.1975
It is estimated that more than 4 billion people worldwide have been infected with hepatitis B virus (HBV)[1]. The infection is persistent in over 350 million individuals with 20%-30% risk of death from HBV-related liver failure or liver cirrhosis[2]. HBV has a circular and partial double-strand DNA genome of 3.2 kb containing four overlapping open reading frames[3]. Based on nucleotide differences of 8% or more in the complete genome, HBV has been classified into eight genotypes from A to H[4-6]. HBV genotype C (HBV/C) induces more severe liver diseases, including hepatocellular carcinoma (HCC), than HBV/B in Asia[7]. Furthermore, patients with HBV genotypes C and D have a lower response rate to interferon alpha (IFN-α) compared with genotypes A and B[8]. Therefore, the analysis of HBV genotype infecting a patient may assist clinical and therapeutic decisions.
Although many HBV genotyping methods exist, there is no standardized or commercially available method for direct molecular typing of the HBV genome. An HBV genotype assay based on the oligonucleotide chip was developed for standardization and potential diagnostic use. This method is based on the reverse hybridization principle so that Cy5-labeled amplicons hybridize to genotype-specific oligonucleotide probes that are immobilized on slides.
In Eastern China, HBV infection is highly prevalent and is an important cause of liver diseases. However, the HBV genotype distribution of the whole area in Eastern China has not been reported systemically. In this study, 400 HBV DNA-positive serum samples collected from patients of the eight cities, including Nanjing, Zhengjiang, Changzhou, Xuzhou, Shanghai, Ningbo, Pingxiang, and Suzhou, in Eastern China were tested using the oligonucleotide chip. In addition, part of results was compared with sequencing.
A total of 400 HBV DNA-positive sera were collected from patients of the eight cities, including Nanjing, Zhengjiang, Changzhou, Xuzhou, Shanghai, Ningbo, Pingxiang, and Hangzhou, in Eastern China from October 2004 to May 2006. Serum samples were taken from 44 asymptomatic HBV carriers (ASC) and 356 subjects with liver disease, consisting of 18 acute hepatitis (AH), 251 chronic hepatitis (CH), 74 liver cirrhosis (LC) and 13 hepatocellular carcinoma (HCC) patients. ASC usually had normal liver function test and no physical signs and symptoms. CH had chronic inflammatory reaction continuously. LC and HCC were diagnosed by ultrasonography and alpha-fetoprotein (AFP) level. All samples came from inpatient and all were known to have positive surface antigen (HbsAg) for more than 6 mo. The demographic data of the patients are listed in Table 1. Sera were tested for ALT, HbsAg, HbsAb, HbeAg, HbeAb, HbcAb using the related kits (Shanghai Kehua Bio-engineering Co. Ltd, Shanghai, China). HBV DNA was quantified using the kit produced by Shenzhen PG Biotech Co. Ltd. The sera were stored at -80°C until analysis.
| Gender (male/female) | Age (yr) | HBV DNA (copies/mL) | HbeAg+/HbeAg - | ALT (IU/L) | |
| Acute infection (n = 18) | 13/5 | 33.1 ± 9.8 | 1.5 × 107 (3.6 × 104 - 9.6 × 109) | 14/4 | 317.1 ± 630.4 |
| Asymptomatic carrier (n = 44) | 27/17 | 28.7 ± 10.2 | 4.6 × 104 (1.2 × 103 - 5.6 × 106) | 14/30 | 15.4 ± 8.7 |
| Chronic hepatitis (n = 251) | 190/61 | 39.1 ± 10.1 | 1.4 × 106 (1.3 × 103 - 7.5 × 109) | 165/86 | 219.6 ± 312.4 |
| Liver cirrhosis (n = 74) | 47/27 | 49.7 ± 16.2 | 2.2 × 105 (5.3 × 103 - 4.7 × 106) | 33/41 | 49.3 ± 59.8 |
| HCC (n = 13) | 10/3 | 54.3 ± 15.4 | 5.6 × 104 (1.2 × 103 - 3.8 × 106) | 7/6 | 192 ± 327.1 |
Viral DNA was extracted from 200 μL of serum samples by using a QIAamp DNA Blood Mini kit (Qiagen Inc., Germany). The target DNA for hybridization was amplified by a nested PCR. A 418-bp product was amplified with the outer primers (sense primer, 5’-ACGYAGCGCCTCATTTTGTG-3’; antisense primer, 5’-CACTGCATGGCCTGAGGAT-3’) which were specific for the pre-S region of HBV. The second stage amplification generated 270-bp product with inner primers (sense primer, 5’-GGGTCACCATATTCTTGGGAA-3’; antisense primer, 5’-Cy5-TGAGGGCTCCACCCCA-3’). Each PCR mixture (40 μL) contained 4 μL of template DNA, 1 × PCR buffer, 1.5 mmol MgCl2, 200 μmol/L of each deoxynucleoside triphosphate, 20 pmol of each primer, and 2 U of Taq DNA polymerase (all primers and reagents were supplied by TaKaRa Biotechnology Inc., Dalian, China). Both two round PCR were performed with a thermocycle (PCR Express; Thermohybaid, UK), and the samples were amplified through 35 cycles, each amplification cycle consisting of denaturation at 94°C for 30 s, primer annealing at 55°C for 30 s, and extension at 72°C for 4 min. Cycles were preceded by incubation at 94°C for 4 min to ensure full denaturation of the target gene and were followed by an extra incubation at 72°C for 10 min to ensure full extension of the products.
A total of 128 sequences of whole HBV genome from GenBank providing clear branching of the eight genotypes with A (n = 19), B (n = 21), C (n = 30), D (n = 21), E (n = 12), F (n = 13), G (n = 6), and H (n = 6) were aligned by using DNA-Star software (Lasergene, Madison, Wis). Eight consensuses of different genotypes were obtained by the alignments of each genotype. When comparing the alignment of each genotype with the alignment of eight consensuses, the result showed that there was a consensus sequence specific to each genotype at the same nucleotide positions in pre-S region among different isolates for each genotype. According to this result, genotype-specific probes were designed for different genotypes of HBV with DNA-Star software (Table 2). Based on the most conservative sequence of pre-S region, a positive control probe (PC probe) was designed for HBV DNA detection. To evaluate the quality of oligonucleotide chip fabrication and hybridization, a quality control probe (QC probe) was designed which reversely complemented the inner anti-sense primer. Probes were synthesized by TaKaRa Biotechnology Inc. All probes contained amino linkers at 5’ ends so that they could covalently attach to aldehyde-coated slides (CEL Associates Inc., Texas). A space arm with a poly (T) 16-mer was inserted between the probe sequence and the 5’ amino linker to decrease steric interference on the chips.
| Genotype | Oligonucleotide sequence | Tm (°C) |
| A | 5’-NH2-(T)16-AAC CCC (A/G)TC AAG GAC CAC TG-3’ | 56 |
| B | 5’-NH2-(T)16-CCC TGC ATT CAA AGC CAA CTC-3’ | 56 |
| C | 5’-NH2-(T)16-CTT CAA CCC CAA CAA GGA TCA-3’ | 54 |
| D | 5’-NH2-(T)16-CAA TCC CAA CAA GGA CAC CTG-3’ | 55 |
| E | 5’-NH2-(T)16-GAC CAC AAT CCC AAC AAA GAC C-3’ | 55 |
| F | 5’-NH2-(T)16-TGG CCA ATG GCA AAC AAG G-3’ | 54 |
| G | 5’-NH2-(T)16-CAA TCC CAA AAA GGA CCC TTG-3’ | 54 |
| H | 5’-NH2-(T)16-CTC TCA ACG GCG AGA AGG G-3’ | 54 |
| QC probe | 5’-NH2-(T)16-TGG GGT GGA GCC CTC AG-3’ | 59 |
| PC probe | 5’-NH2-(T)16- ATC CTC AGG CCA TGC AGT G-3’ | 58 |
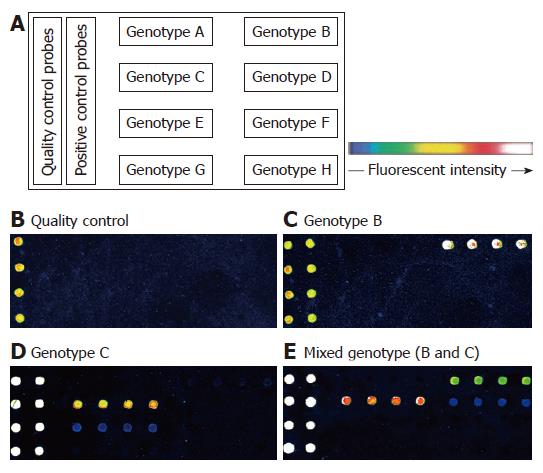
The probes were diluted to a final concentration of 10 μmol/L with spotting solution (TeleChem, USA) and spotted onto aldehyde-coated slides with a robot (Cartesian Pixsys 7500, USA). Four replicate spots of each probe were aligned in the same rows. QC and PC probes were spotted on the first and second left column on the chip (Figure 1). After the spotting process, the oligonucleotide chips were fixed at 25°C for 48 h and dried for further use.
Two microliters of Cy5-labeled unpurified PCR product was mixed with 10 μL of DIG Easy Hyb (Roche Molecular Biochemicals, Canada). The mixture was denatured for 2 min at 95°C and then chilled on ice for 5 min. The hybridization mixture was applied to the array and covered with a glass cover slip to prevent evaporation of the mixture during the hybridization. The oligonucleotide chips were incubated at 40°C for 30 min in a moist incubator. After hybridization, the washing procedure was carried out as described in our previous research[9]. The oligonucleotide chip images were obtained with a fluorescent scanner (GenePix 4000B; Axon Instruments, Inc., Calif).
The products of the first-round PCR were monitored on agarose gel to ensure that sufficient DNA was available for sequencing. Eighty products were randomly selected for the phylogenetic analysis. The outer sense primer also was used for DNA sequencing. The sequence was determined by Shanghai Bioasia Biotechnology Ltd. with ABI Prism 3730 genetic analyzer (Applied Biosystems, Inc.). The sequence datum of each selected specimen was aligned with pre-S sequences of eight consensuses for different genotypes which were obtained from the alignment of 128 complete HBV genome sequences in GenBank. Alignment and phylogenetic analysis were performed with DNA-Star software by using Clustal W.
Two HBV genotypes (B, C) and a mixed type (B and C) were detected among the 400 samples using our oligonucleotide chips. The layout of oligonucleotide chip and the representative fluorescent scanning of genotyping detection are shown in Figure 1. Genotype distribution in Eastern China in our study was finally determined as follows: genotype C, 333 (83.2%); genotype B, 33 (8.3%); and mixed genotype (B and C), 34 (8.5%). The other genotypes were not detected in this study.
Genotype of the HBV DNA in 80 specimens was performed by sequencing and phylogenetic analysis of the pre-S region, and these genotypes were compared to the genotypes determined by the oligonucleotide chip for the same specimens. Phylogenetic analysis of pre-S sequence for each specimen by using the eight consensus sequences of different genotypes for comparison provided clear genotyping determination for each sample (data not shown). The results of the two methods for single genotypes were 100% concordant. The oligonucleotide chip detected nine mixed genotypes which were determined by sequencing in a single genotype. Four of them were selected for clonal analysis. The PCR products of the four samples were ligated into pGEM-T easy vector (Promega), then the ligated products were transformed into E. coli DH5α (Clontech). Individual colonies were tested by the oligonucleotide chip at first, and then were sent to be sequenced. Clonal analysis confirmed that all the four specimens were mixed genotypes.
There are eight genotypes of HBV designated A to H based on greater than 8% nucleotide variation over the entire genome. The eight genotypes of HBV show a distinct geographical distribution and influence the course of disease and the efficacy of treatment. Genotype A is prevalent in Northern and Central Europe, but is also common in North America and sub-Saharan Africa. Genotypes B and C are confined to Asia, genotype D is widespread but is the predominant genotype in the Mediterranean region, while genotype E is found mainly in West Africa[10]. Genotype F shows the highest divergence among the genotypes and is indigenous to aboriginal populations of the Americas[11]. Genotype G is found in USA, France, and Germany[5,12,13], genotype H in the Central America[6]. Thus, the variability of HBV genotypes can influence the interpretation of diagnostic data and the therapeutic decisions.
Our results showed that genotype B, C and mixed genotype exist in Eastern China. Genotype C is the major genotype in this area (accounting for 83.2%), which is similar with HBV genotype distribution in China in previous reports[7,14-16]. Genotype B was more frequently found in patients with mild hepatic inflammation (ASC), while genotype C in those with advanced liver diseases (CH and LC) (P < 0.05) (Table 3). Although there was no significant difference, infection with genotype C showed highest frequency (92.3%) in the patients with HCC. This result may correlate with the higher frequent mutations in the BCP of genotype C compared with to genotype B[8]. Meanwhile the major risk for liver cirrhosis is exacerbated by the acquisition of basic core promoter (BCP) mutations that accelerate carcinogenesis[17]. There were no significant differences in the median of HBV DNA and ALT levels between the patients with genotypes B and C.
In this study, immobilization of oligonucleotide probes onto an aldehyde-coated glass surface produced a high-density monolayer that was easily accessible for hybridization with DNA samples, which made the reaction between the fluorecence-labeled DNA targets and oligonucleotide probes more efficient and quick, and reduced the overall assay time to less than 5 h. Due to the high degree of allele specificity of the oligonucleotide probes for the pre-S sequence and the highly stringent conditions that were used, only the target specifically complementary to the probe could be detected. To increase the hybridization efficiency between the probes and targets and sensitivity of the assay and to obtain higher signal-to-noise ratios, the lengths of the probes and spacer arms and the hybridization condition were optimized. Oligonucleotide chip assay is based on the sequence of pre-S region which has been used for genotyping purposes shown to provide genotypic determinations similar to those provided by full genome analysis. Our results demonstrated a high degree of correlation between the genotypes determined by sequencing of the pre-S region and oligonucleotide chip assay. Discrepancies between the two methods appear to be due to the presence of mixed genotype infections. We detected 34 samples with genotypic co-infections by oligonucleotide chips, four of these samples were further analyzed, genotypic co-infections were confirmed by clonal analysis. Thus, our results suggest that oligonucleotide chip assay has a very high sensitivity for the detection of multiple genotypes.
Since Okamoto et al[18] established the first method to classify HBV, several methods for HBV genotyping have been reported except sequence analysis, such as multiplex PCR[19], restriction fragment length polymorphism (RFLP)[20], serological assay[21] and reverse hybridization[22]. These other genotyping methods have a number of advantages over sequence analysis as they are inexpensive and easy to perform. The major disadvantage is that only a limited number of nucleotides/amino acids are actually analyzed and therefore these methods are not as reliable as sequence plus phylogenetic analysis. In addition, these genotyping methods cannot distinguish the new genotypes, such as HBV genotype G and H. Thus, a better sequence-specific approach is needed. The development of a rapid, sensitive, and accurate method for the identification of HBV genotyping is therefore valuable and reasonable. Our oligonucleotide chip assay is designed to detect all eight genotypes of HBV according to the results of a large scale of phylogenetic analysis.
In conclusion, the present study shows that genotype C and B are the two common HBV genotypes in Eastern China. Genotype C is the major genotype in this area. Meanwhile the oligonucleotide chip assay is a promising tool for HBV genotyping.
This article is mainly about a new method for the detection of HBV genotyping. There are eight genotypes of HBV designated A to H based on greater than 8% nucleotide variation over the entire genome. The eight genotypes of HBV showed a distinct geographical distribution and influence the course of disease and the efficacy of treatment.
Since Okamoto et al[18] established the first method to classify HBV, many methods for HBV genotyping have been developed, such as multiplex PCR, RFLP, serological assay, and reverse hybridization. More and more studies have been dedicated to revealing the influence of HBV genotype on the clinical prognosis and antiviral therapy.
Many methods for HBV genotyping, such as sequence analysis, RFLP, PCR, line probe assay, have been developed. Line probe assay is the most sensitive and accurate. However, it is very laborious and expensive. As the oligonucleotide probes are immobilized on the slides in our method, the non-specific hybridization will be reduced greatly. So the protocol of hybridization can be simplified and whole test can be finished in 5 h. Thus, this study provides a rapid, sensitive, convenient, accurate and affordable method for the identification of HBV genotyping.
This study provides a new method for clinical diagnosis of HBV genotyping which would influence the efficacy of treatment.
Oligonucleotide chip is manufactured by using a set of sequence-specific oligonucleotide probes derived from polymorphic regions of requested genes. Phylogenetic analysis is the process to determine the evolutionary relationships between organisms. The results of an analysis can be drawn in a hierarchical diagram called a phylogenetic tree. Originally, phylogenetic trees were created using morphology, but now, determining evolutionary relationships includes matching patterns in nucleic acid and protein sequences.
This article provides a new HBV genotype assay based on the oligonucleotide chip. This method is based on the reverse hybridization principle, so that Cy5-labeled amplicons hybridize to genotype-specific oligonucleotide probes that are immobilized on slides. As mentioned in the article, 400 HBV DNA-positive serum samples collected from patients in Eastern China were tested using the oligonucleotide chip. The results showed that genotypes C and B were the two common HBV genotypes in Eastern China and genotype C is the major genotype in this area. Meanwhile the oligonucleotide chip assay is a promising tool for HBV genotyping.
S- Editor Liu Y L- Editor Kumar M E- Editor Ma WH
| 1. | Lai CL, Ratziu V, Yuen MF, Poynard T. Viral hepatitis B. Lancet. 2003;362:2089-2094. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 595] [Cited by in F6Publishing: 582] [Article Influence: 27.7] [Reference Citation Analysis (0)] |
| 2. | Kao JH, Chen DS. Global control of hepatitis B virus infection. Lancet Infect Dis. 2002;2:395-403. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 596] [Cited by in F6Publishing: 595] [Article Influence: 27.0] [Reference Citation Analysis (0)] |
| 3. | Magnius LO, Norder H. Subtypes, genotypes and molecular epidemiology of the hepatitis B virus as reflected by sequence variability of the S-gene. Intervirology. 1995;38:24-34. [PubMed] [Cited in This Article: ] |
| 4. | Norder H, Couroucé AM, Magnius LO. Complete genomes, phylogenetic relatedness, and structural proteins of six strains of the hepatitis B virus, four of which represent two new genotypes. Virology. 1994;198:489-503. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 556] [Cited by in F6Publishing: 599] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 5. | Stuyver L, De Gendt S, Van Geyt C, Zoulim F, Fried M, Schinazi RF, Rossau R. A new genotype of hepatitis B virus: complete genome and phylogenetic relatedness. J Gen Virol. 2000;81:67-74. [PubMed] [Cited in This Article: ] |
| 6. | Arauz-Ruiz P, Norder H, Robertson BH, Magnius LO. Genotype H: a new Amerindian genotype of hepatitis B virus revealed in Central America. J Gen Virol. 2002;83:2059-2073. [PubMed] [Cited in This Article: ] |
| 7. | Ding X, Mizokami M, Ge X, Orito E, Iino S, Ueda R, Nakanishi M. Different hepatitis B virus genotype distributions among asymptomatic carriers and patients with liver diseases in Nanning, southern China. Hepatol Res. 2002;22:37-44. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 34] [Cited by in F6Publishing: 40] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 8. | Kao JH, Chen PJ, Lai MY, Chen DS. Hepatitis B genotypes correlate with clinical outcomes in patients with chronic hepatitis B. Gastroenterology. 2000;118:554-559. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 684] [Cited by in F6Publishing: 668] [Article Influence: 27.8] [Reference Citation Analysis (0)] |
| 9. | Zhou W, Du W, Cao H, Zhao J, Yang S, Li W, Shen Y, Zhang S, Du W, Zhang X. Detection of gyrA and parC mutations associated with ciprofloxacin resistance in Neisseria gonorrhoeae by use of oligonucleotide biochip technology. J Clin Microbiol. 2004;42:5819-5824. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 15] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 10. | Norder H, Hammas B, Lee SD, Bile K, Couroucé AM, Mushahwar IK, Magnius LO. Genetic relatedness of hepatitis B viral strains of diverse geographical origin and natural variations in the primary structure of the surface antigen. J Gen Virol. 1993;74:1341-1348. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 247] [Cited by in F6Publishing: 266] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 11. | Sánchez LV, Maldonado M, Bastidas-Ramírez BE, Norder H, Panduro A. Genotypes and S-gene variability of Mexican hepatitis B virus strains. J Med Virol. 2002;68:24-32. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 63] [Cited by in F6Publishing: 67] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 12. | Kato H, Orito E, Gish RG, Sugauchi F, Suzuki S, Ueda R, Miyakawa Y, Mizokami M. Characteristics of hepatitis B virus isolates of genotype G and their phylogenetic differences from the other six genotypes (A through F). J Virol. 2002;76:6131-6137. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 97] [Cited by in F6Publishing: 105] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 13. | Vieth S, Manegold C, Drosten C, Nippraschk T, Günther S. Sequence and phylogenetic analysis of hepatitis B virus genotype G isolated in Germany. Virus Genes. 2002;24:153-156. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 61] [Cited by in F6Publishing: 67] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 14. | Ding X, Mizokami M, Yao G, Xu B, Orito E, Ueda R, Nakanishi M. Hepatitis B virus genotype distribution among chronic hepatitis B virus carriers in Shanghai, China. Intervirology. 2001;44:43-47. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 131] [Cited by in F6Publishing: 144] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 15. | Yuen MF, Sablon E, Tanaka Y, Kato T, Mizokami M, Doutreloigne J, Yuan HJ, Wong DK, Sum SM, Lai CL. Epidemiological study of hepatitis B virus genotypes, core promoter and precore mutations of chronic hepatitis B infection in Hong Kong. J Hepatol. 2004;41:119-125. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 49] [Cited by in F6Publishing: 53] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 16. | Fang ZL, Zhuang H, Wang XY, Ge XM, Harrison TJ. Hepatitis B virus genotypes, phylogeny and occult infection in a region with a high incidence of hepatocellular carcinoma in China. World J Gastroenterol. 2004;10:3264-3268. [PubMed] [Cited in This Article: ] |
| 17. | Kuang SY, Jackson PE, Wang JB, Lu PX, Muñoz A, Qian GS, Kensler TW, Groopman JD. Specific mutations of hepatitis B virus in plasma predict liver cancer development. Proc Natl Acad Sci USA. 2004;101:3575-3580. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 128] [Cited by in F6Publishing: 139] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 18. | Okamoto H, Tsuda F, Sakugawa H, Sastrosoewignjo RI, Imai M, Miyakawa Y, Mayumi M. Typing hepatitis B virus by homology in nucleotide sequence: comparison of surface antigen subtypes. J Gen Virol. 1988;69:2575-2583. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 725] [Cited by in F6Publishing: 750] [Article Influence: 20.8] [Reference Citation Analysis (0)] |
| 19. | Mizokami M, Nakano T, Orito E, Tanaka Y, Sakugawa H, Mukaide M, Robertson BH. Hepatitis B virus genotype assignment using restriction fragment length polymorphism patterns. FEBS Lett. 1999;450:66-71. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 183] [Cited by in F6Publishing: 180] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 20. | Naito H, Hayashi S, Abe K. Rapid and specific genotyping system for hepatitis B virus corresponding to six major genotypes by PCR using type-specific primers. J Clin Microbiol. 2001;39:362-364. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 184] [Cited by in F6Publishing: 210] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 21. | Usuda S, Okamoto H, Tanaka T, Kidd-Ljunggren K, Holland PV, Miyakawa Y, Mayumi M. Differentiation of hepatitis B virus genotypes D and E by ELISA using monoclonal antibodies to epitopes on the preS2-region product. J Virol Methods. 2000;87:81-89. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 64] [Cited by in F6Publishing: 67] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 22. | Osiowy C, Giles E. Evaluation of the INNO-LiPA HBV genotyping assay for determination of hepatitis B virus genotype. J Clin Microbiol. 2003;41:5473-5477. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 87] [Cited by in F6Publishing: 96] [Article Influence: 4.8] [Reference Citation Analysis (0)] |