Published online Mar 14, 2007. doi: 10.3748/wjg.v13.i10.1602
Revised: February 3, 2006
Accepted: February 27, 2007
Published online: March 14, 2007
AIM: To clone and identify human genes transactivated by PS1TP5 by constructing a cDNA subtractive library with suppression subtractive hybridization (SSH) technique.
METHODS: SSH and bioinformatics techniques were used for screening and cloning of the target genes transactivated by PS1TP5 protein. The mRNA was isolated from HepG2 cells transfected with pcDNA3.1(-)-myc-his(A)-PS1TP5 and pcDNA3.1(-)-myc-his(A) empty vector, respectively, and SSH technique was employed to analyze the differentially expressed DNA sequence between the two groups. After digestion with restriction enzyme RsaI, small size cDNAs were obtained. Then tester cDNA was divided into two groups and ligated to the specific adaptor 1 and adaptor 2, respectively. The tester cDNA was hybridized with driver cDNA twice and subjected to nested PCR for two times, and then subcloned into T/A plasmid vectors to set up the subtractive library. Amplification of the library was carried out with E. coli strain DH5α. The cDNA was sequenced and analyzed in GenBank with Vector NTI 9.1 and NCBI BLAST software after PCR amplification.
RESULTS: The subtractive library of genes transactivated by PS1TP5 was constructed successfully. The amplified library contained 90 positive clones. Colony PCR showed that 70 clones contained 200-1000-bp inserts. Sequence analysis was performed in 30 clones randomly, and the full-length sequences were obtained by bioinformatics technique. Altogether 24 coding sequences were obtained, which consisted of 23 known and 1 unknown. One novel gene with unknown functions was found and named as PS1TP5TP1 after being electronically spliced, and deposited in GenBank (accession number: DQ487761).
CONCLUSION: PS1TP5 is closely correlated with immunoregulation, carbohydrate metabolism, signal transduction, formation mechanism of hepatic fibrosis, and occurrence and development of tumor. Understanding PS1TP5 transactive proteins may help to bring some new clues for further studying the biological functions of pre-S1 protein.
- Citation: Zhang JK, Zhao LF, Cheng J, Guo J, Wang DQ, Hong Y, Mao Y. Screening and cloning for proteins transactivated by the PS1TP5 protein of hepatitis B virus: A suppression subtractive hybridization study. World J Gastroenterol 2007; 13(10): 1602-1607
- URL: https://www.wjgnet.com/1007-9327/full/v13/i10/1602.htm
- DOI: https://dx.doi.org/10.3748/wjg.v13.i10.1602
Hepatitis B virus (HBV) causes acute and chronic infections of the liver. Acute infections may result in serious disease with approximately 0.5% of cases developing into fatal, fulminant hepatitis. Chronic infections may also have remarkable consequences. In addition to causing acute and chronic hepatitis, HBV is considered a major etiological factor in the development of human hepatocellular carcinoma (HCC), one of the most frequent fatal malignancies worldwide and worldwide deaths of HCC exceed one million per year. Epidemiological studies have demonstrated an approximately 10-fold increase in the relative risk of HCC among HBV carriers compared to non-carriers[1-3].
The precise role of HBV in the etiology of HCC is not well understood. Only occasionally are genes controlling cell growth and differentiation disturbed by integration of HBV DNA sequences. An alternative mechanism of chronic infection and hepatocarcinogenesis may be the key steps to mutual interaction between viral proteins and hepatocellular proteins. This action may mediate the virus to enter into the liver cells and affect the activities and function of these proteins. Moreover, proteins from hepatocytes infected with HBV inversely disturb virus replication and reduce immunity of the host, resulting in chronic liver diseases and HCC. Understanding of the interaction among these proteins may help to bring some new clues for discovering the pathogenesis of viral hepatitis and related HCC.
The first full-length nucleotide sequence of HBV was published in 1979[4]. The four open reading frames (ORF) defined in the HBV genome at that time were named as the regions of S, C, P and X. The region of S was divided into the sub-regions of pre-S1, pre-S2 and S according to different initial code ATG in frame. Dong et al[5] have shown that there is an ORF before the pre-S1 region in the genome of HBV and they amplified it from the serum of patients infected with HBV by long and accurate polymerase chain reaction (LAPCR). This region is of 135 bp and tentatively named the pre-pre-S and its promoter activity has been confirmed in a 277-bp upstream nucleotide sequence before the pre-S1 gene[6]. Thus, the complete S region of HBV includes pre-pre-S, pre-S1, pre-S2 and S subregions.
The function of the pre-S1 protein in the life cycle of HBV remains unknown. Its transactive function has been demonstrated by some recent studies[7,8]. Human gene 5 transactivated by pre-S1 protein of HBV (PS1TP5) is a novel target gene that has been screened and cloned with a suppression subtractive hybridization (SSH) technique in our laboratory (GenBank accession number: AY427953)[9]. The full length of the coding rank of PS1TP5 contains 438 nucleotides, and the protein product consists of 145 amino acid residues. To further investigate the biological significance of the PS1TP5 and pre-S1 protein, we screened and identified the proteins transactivated by PS1TP5 protein using the SSH technique.
HepG2 cell line of hepatoblastoma and E. coli strains DH5α were conserved in our laboratory. Eukaryotic expression vector pcDNA3.1(-)-myc-his(A), Quickprep mico mRNA Purification kit, PCR-Select cDNA Subtraction kit, 50 × PCR Enzyme Mix kit and Advantage PCR Cloning kit were purchased from Clontech Co., America. FuGENE6 transfection kit was purchased from Roche Co., America. pGEM-T vector, pGEM-T-easy vector and High Pure PCR Product Purification kit were from Promega Co., America. EcoRI, BgIII and DNA Marker were from Takara Company, Japan. DNA sequencing was performed by Invitrogen Company, China.
The reverse transcription polymerase chain reaction (RT-PCR) was performed to amplify the gene of PS1TP5 from the mRNA of HepG2 cells. The sequences of the primers containing the EcoRI and BgIII restriction enzyme sites were: 5’-CGG AAT TCA TGG GCT TGA AGA GCC AC-3’ (sense) and 5’-CGA GAT CTA GTG AAG ATA TGC AGA GG-3’ (anti-sense). Samples were amplified through 35 cycles, each amplification cycle consisting of denaturation at 94°C for 45 s, primer annealing at 60°C for 45 s, and extension at 72°C for 1 min. Ten nanograms of the 438-bp PCR product was cloned into pGEM-T vector. The primary structure of insert was confirmed by direct sequencing. The fragment of encoding PS1TP5 was released from the pEGM-T-PS1TP5 by digestion with EcoRI and BgIII, and ligated to the EcoRI/BgIII sites of pcDNA3.1(-)-myc-his(A) empty vector. Recombinant eukaryotic expression vector pcDNA3.1(-)-myc-his(A)-PS1TP5 was obtained, then identified by PCR and digested with EcoRI/BgIII.
HepG2 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 100 kU/L penicillin, 100 mg/L streptomycin, and 100 mL/L heat-inactivated fetal bovine serum (FBS), and incubated at 37°C in a humidified atmosphere consisting of 50 mL/L CO2 in air. About 1.5 × 106 cells were seeded in 35-mm plates 12 h prior to transfection, which reached 50% confluence at the time of transfection. Cells were transfected with FuGENE6 transfection reagent using 2.0 μg of pcDNA3.1(-)-myc-his(A) empty (driver) and pcDNA3.1(-)-myc-his(A)-PS1TP5 plasmid DNA (tester) according to the manufacturer’s protocol.
The mRNA from HepG2 cells transfected with pcDNA3.1(-)-myc-his(A)-PS1TP5 (tester) and pcDNA3.1(-)-myc-his(A) empty vector (driver) was isolated using a micro mRNA Purification kit, respectively, and cDNAs were reverse-transcribed from total RNA. Quantitative analysis of mRNA was carried out with an ultraviolet spectrophotometer. Identification was done by PCR with PS1TP5 sequence-specific primers.
Genome comparison was performed by suppression subtraction hybridization (SSH) technique according to the manufacturer’s instructions of PCR-select cDNA subtraction kit. Briefly, 2 μg of mRNA from the tester and the driver was subjected to cDNA synthesis. Tester and driver cDNAs were digested with RsaI. The tester cDNA was split into two groups, and each was ligated with a different cDNA adapter. In the first hybridization reaction, an excess of driver was added to each sample of the tester. The samples were heat-denaturated and allowed to anneal. Because of the second-order kinetics of hybridization, the concentration of high- and low-abundance sequences is equalized among the single-stranded tester molecules. At the same time, single-stranded tester molecules were significantly enriched for differentially expressed sequences. During the second hybridization, the two primary hybridization samples were mixed together without denaturation. Only the remaining equalized and subtracted single-stranded tester cDNAs can re-associate forming double-stranded tester molecules with different ends. After filling in the ends with DNA polymerase, the entire population of molecules was subjected to nested PCR with two adapter-specific primer pairs. Then secondary PCR products were used as templates for PCR amplification of G3PDH (a housekeeping gene) at 18, 23, 28, 33 cycles to assure subtracted efficiency.
Products of these amplified overhangs containing a subtracted cDNA library (6 μL) were ligated into a pGEM-T-easy vector. Subsequently, the plasmid was transformed into E. coli strain DH5α. Bacteria were cultured in 800 μL of LB medium and allowed to incubate for 45 min at 37°C and 225 r/min. After incubation, bacteria were plated onto agar plates containing ampicillin (100 mg/L), 5-bromo-4-chloro-3-indolyl-β-D-galactoside (X-gal, 20 μg/cm2) and isopropyl-β-D-thiogalactoside (IPTG, 12.1 μg/cm2) and incubated overnight at 37°C. White colonies were selected and identified by PCR. Primers were T7/SP6 primer of pGEM-T-easy vector. After sequencing the plasmids DNA of positive colonies, nucleic acid homology searches were performed using Vector NTI 9.1 and NCBI BLAST software.
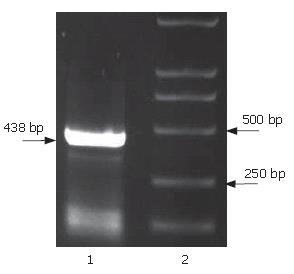
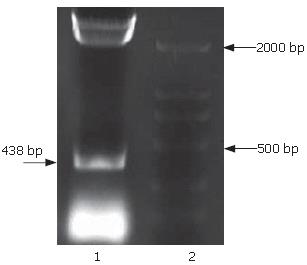
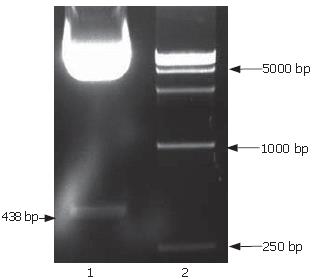
The full-length sequences of PS1TP5 were generated by RT-PCR amplification of the mRNA of HepG2 cells, subcloned into pGEM-T vector, analyzed restrictively with EcoRI/BgIII and sequenced by comparing to vector NTI 9.1 and conducting a BLAST database homology search. Analysis of the PS1TP5 PCR reaction products (Figure 1) and recombinant plasmid pGEM-T-PS1TP5 (Figure 2) by agarose gel electrophoresis showed the clear bands with the expected size of 438 bp. Sequences of the PCR products were corrected using bioinformatic analysis. After being cut from pGEM-T-PS1TP5 by EcoRI/BgIII, the fragment was ligated in-frame into a pcDNA3.1(-)-myc-his(A) EcoRI/BgIII site. Restriction enzyme analysis of the pcDNA3.1(-)-myc-his(A)-PS1TP5 plasmid with EcoRI/BgIII yielded two bands: approximately 5500-bp empty pcDNA3.1(-)-myc-his(A); and a 438-bp PS1TP5 (Figure 3).
The mRNA isolated from HepG2 cells transfected with pcDNA3.1(-)-myc-his(A)-PS1TP5 (tester) and pcDNA3.1(-)-myc-his(A) empty vector (driver) was detected using an ultraviolet spectrophotometer (Table 1). Quantitative results of mRNA suggested that isolated mRNA could be used for suppression subtractive hybridization.
| Transfectedplasmid | A230 | A260 | A280 | A260/A280 | mRNAquantitation(mg/L) |
| pcDNA3.1(-)-myc- his(A)-PS1TP5 | 0.173A | 0.377A | 0.172A | 2.192 | 15.08 |
| pcDNA3.1(-)-myc- his(A) | 0.191A | 0.378A | 0.173A | 2.185 | 14.84 |
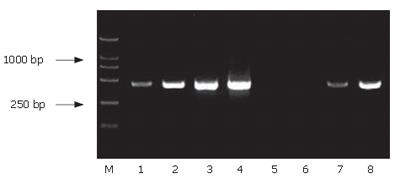
The G3PDH (a housekeeping gene) primers were used to confirm the reduced relative abundance of G3PDH following the PCR selection procedure. The result displayed that the G3PDH abundance of the subtracted secondary PCR products significantly decreased compared to the unsubtracted one, indicating that the subtractive library had a high subtraction efficiency (Figure 4).
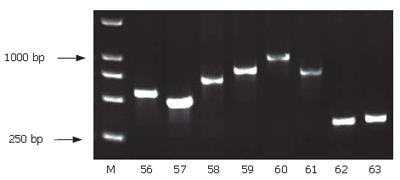
Using SSH technique, we obtained a total of 90 positive clones. These clones were prescreened by PCR amplification to ensure that only clones with different inserts were subjected to sequencing (Figure 5). Among these clones, 70 clones contained 200-1000-bp inserts. A total of 30 clones from the cDNA library were randomly chosen and sequenced. Using the BLAST program at the National Center for Biotechnology Information, 23 sequences from 29 true positive colonies had a high similarity to known genes and one sequence failed to match with a homologous gene in GenBank (Table 2).
| High similarity proteins to known genes | n | Homology (%) |
| Homo sapiens ribosomal protein S16 (RPS16) | 2 | 98 |
| Homo sapiens ribosomal protein LP0 (RPLP0) | 1 | 98 |
| Homo sapiens ribosomal protein LP2 (RPLP2) | 1 | 100 |
| Homo sapiens ribosomal protein S7 (RPS7) | 1 | 100 |
| Homo sapiens ribosomal protein L31 (RPL31) | 1 | 100 |
| Homo sapiens transmembrane 4 superfamily member 4-CD81 (TM4SF4) | 1 | 99 |
| Homo sapiens inositol monophosphatase 2 (IMPA2) | 2 | 99 |
| Homo sapiens protein kinase BRPK | 1 | 100 |
| Homo sapiens clusterin (CLU) | 1 | 100 |
| Homo sapiens adenosine deaminase | 2 | 100 |
| Homo sapiens solute carrier family 7 member 5 (SLC7A5) | 1 | 99 |
| Homo sapiens cytochrome c oxidase subunit 8A | 2 | 100 |
| Homo sapiens enolase 1 | 1 | 99 |
| Homo sapiens H3 histone family 3B | 1 | 100 |
| Homo sapiens replication factor C | 2 | 98 |
| Human cytoskeletal gamma-actin gene | 1 | 100 |
| Homo sapiens S100 calcium-binding protein A6 (calcyclin) | 1 | 99 |
| Homo sapiens eukaryotic translation elongation factor 1 | 1 | 100 |
| Homo sapiens epoxide hydrolase 1 | 1 | 98 |
| Homo sapiens dehydrogenase 1 | 1 | 99 |
| Homo sapiens signal sequence receptor-β | 2 | 100 |
| Homo sapiens β5-microtubulin | 1 | 99 |
| Homo sapiens pyruvate dehydrogenase | 1 | 98 |
| New genes with unknown function | 1 | 100 |
According to Kozak regulation of start codon and signal sequences of downstream conservative poly adenine, one sequence with unknown function was electronically analyzed, spliced, and concluded with Vector NTI 9.1 and NCBI BLAST software. A new gene was obtained and named PS1TP5TP1. The full length of the coding rank of PS1TP5TP1 contained 237 nucleotides, and the coding product consisted of 78 amino acid residues (GenBank accession number: DQ487761, Figure 6).
The open reading frame (ORF) of the HBV complete S gene consists of four coding regions: pre-pre-S, pre-S1, pre-S2 and S, each starting with an ATG codon in frame. Through in-frame translational initiation at each of the four ATG codons, complete S (pre-pre-S + pre-S1 + pre-S2 + S), large (LHBs; pre-S1 + pre-S2 + S), middle (MHBs; pre-S2 + S) and small (SHBs; S) envelope glycoproteins can be synthesized[10,11]. Interaction between viral and hepatocellular proteins plays an important role in the pathogenesis of the virus and may mediate virus to enter into hepatocytes. Their network interaction can change normal biological function of proteins, influence self-replication of the virus, and result in disease.
Suppression subtractive hybridization (SSH) is a new and highly effective gene analysis method designed by Diatchenko et al[12,13], and has been developed for the generation of subtracted cDNA libraries. It is based primarily on a technique called suppression PCR, and combines normalization and subtraction in a single procedure. The normalization step equalizes the abundance of cDNAs within the target population and the subtraction step excludes the common sequences between the target and driver populations. As a result only one round of subtractive hybridization is needed and the subtracted library is normalized in terms of abundance of different cDNAs. It dramatically increases the probability of obtaining low-abundance differentially expressed cDNA and simplifies analysis of the subtracted library. The SSH technique is applicable to many molecular genetic and positional cloning studies for the identification of disease, developmental, tissue-specific, or other differentially expressed genes.
In this study, we cotransfected HepG2 cells with pcDNA3.1(-)-myc-his(A)-PS1TP5 (tester) and pcDNA3.1(-)-myc-his(A) empty vector (driver). The mRNA was isolated from transfected HepG2 cells, and total RNA was reverse-transcribed into cDNAs. The subtractive library of genes transactivated by PS1TP5 was set up successfully, and 30 clones from the cDNA library were selected randomly and sequenced, and the full-length sequences were obtained with bioinformatics method. By sequence analysis using Vector NTI 9.1 and NCBI BLAST software, we obtained the sequences of the 23 genes with known functions and one gene with unknown functions. After electronic splicing, this new gene with unknown function was named PS1TP5TP1 and deposited in GenBank (accession number: DQ487761).
The 23 genes with known functions are closely correlated with immunoregulation, carbohydrate metabolism, signal transduction, formation of hepatic fibrosis, and initiation and development of tumor. Homo sapiens protein kinase BRPK, a novel protein kinase, is a kind of phosphorylation. BRPK has a serine/threonine-type protein kinase domain, and the recombinant proteins of BRPK are capable of autophosphorylation. Experiments performed by Nagajima et al[14] revealed that BRPK was expressed at a higher level in three carcinoma cell lines with higher metastatic potential, thereby indicating a possible link of BRPK to initiation and development of tumor. Clusterin, also known as apoprotein J, is a kind of glycoprotein heterodimers residing in most mammalian tissue and body fluid. Several studies reported that clusterin was exorbitantly expressed in many kinds of malignant tumors, including breast[15], kidney[16], bladder[17], pancreas[18], colon[19], lymph[20], liver[21], and so on. The formation and expansion of clusterin-nuclear matrix may be related to the activated cell growth. Therefore, it may play an important role in alarm reaction, apoptosis and tumorigenesis. Adenosine deaminase is a nucleic acid metabolic enzyme that is connected with immunological activity of somatocyte, and exists extensively in various kinds of tissues. Some reports suggested that adenosine deaminase deficiency was associated with acquired immunodeficiency syndrome (AIDS) and hepatitis B infection[22,23]. Homo sapiens S100 calcium-binding protein A6 (calcyclin), a cell cycle-regulated protein, is a member of the S100A family of calcium-binding proteins. The calcyclin gene has been localized to the long arm of human chromosome 1. While the precise function of calcyclin was unknown, some experimental observations suggested that the functional role of calcyclin was associated with cell proliferation, signal transduction, pulmonary fibrosis and several types of cancer phenotypes through the cell cycle[24-26]. Homo sapiens replication factor C (RFC) is located in eukaryotic cell. A functional mode of RFC through its interaction with proliferation cell nuclear antigen (PCNA) and cyclin by structural changes is called molecular switch extending DNA. This active characteristic is necessary for not only duplication and recovery of DNA as well as signal checkpoint of cell life, but also intracellular multifunction[21,22]. Mammalian transmembrane 4 superfamily (TM4SF) proteins (also known as tetraspans or tetraspanins) include at least 16 core members and a number of additional proteins with sequence similarities. Almost all mammalian cells contain one or more TM4SF proteins[29]. TM4SF protein CD81 may function in cell migration, proliferation and tumor cell metastasis. Most TM4SF proteins are located at plasma membrane, and several are located in cell lamellipodia and lopodia, consistent with their role in cell motility. TM4SF proteins including CD81 are also found in various intracellular granules and vesicles. A specific subset of TM4SF proteins may recruit PI 4-kinase to specific membrane locations, and thereby influence phosphoinositide-dependent signaling[30]. Ribosomal protein is a group of organella involving in cell structure and cell cycle, and possess the important ability to control cell growth, differentiation and adherence. The ribosomal protein is certainly related to cell energy or substance metabolism, and has critical physiological roles in nutrient transport[31].
HBV PS1TP5 protein also interacts with Homo sapiens inositol monophosphatase 2 (IMPA2), Homo sapiens solute carrier family 7 member 5 (SLC7A5), Homo sapiens cytochrome c oxidase subunit 8A, Homo sapiens enolase 1, Homo sapiens H3 histone family 3B, Human cytoskeletal gamma-actin gene, Homo sapiens eukaryotic translation elongation factor 1, Homo sapiens epoxide hydrolase 1, Homo sapiens dehydrogenase 1, Homo sapiens signal sequence receptor-β, Homo sapiens β5-microtubulin and Homo sapiens pyruvate dehydrogenase. How the interaction between PS1TP5 protein and the aforementioned interacting proteins affects the initiation and development of chronic hepatitis B, hepatic fibrosis and hepatocarcinoma needs to be further studied.
The hepatitis B virus (HBV) genome includes S, C, P and X regions. The S region is divided into four subregions of pre-pre-S, pre-S1, pre-S2 and S. PS1TP5 is a novel target gene transactivated by the pre-S1 protein (GenBank accession number: AY427953) and its precise function in the life cycle of HBV remains unknown. In order to investigate the biological function of the PS1TP5 protein, we performed a suppression subtractive hybridization technique to screen and identify proteins interacting with the PS1TP5 protein by constructing a cDNA subtractive library.
Suppression subtractive hybridization (SSH) is a new and highly effective gene analysis method and is applicable to many molecular genetic and positional cloning studies for differentially expressed genes. Understanding PS1TP5 transactive proteins may help to bring some new clues for discovering pathogenesis of viral hepatitis and related hepatocellular carcinoma.
In this study, we found 23 genes with known functions that were closely correlated with immunoregulation, carbohydrate metabolism, signal transduction, formation of hepatic fibrosis, and initiation and development of tumor. A new gene with unknown function was obtained and named PS1TP5TP1 (GenBank accession number: DQ487761).
Network interaction between viral and hepatocellular proteins plays an important role in the pathogenesis of viral hepatitis and related hepatocellular carcinoma. Studying of PS1TP5 precise role may help to further understand the network interaction and establish foundation for clinical gene therapy.
Trans-activating: Activated mode of gene expression includes cis-activating and trans-activating. The former is intramolecular activating and the latter is intermolecular activating.
Using the suppression subtractive hybridization technique the authors have identified the proteins transactivated by PS1TP5 protein. The study is well conducted and well written.
S- Editor Wang J L- Editor Kumar M E- Editor Lu W
| 1. | Shouval D. Hepatitis B vaccines. J Hepatol. 2003;39 Suppl 1:S70-S76. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 118] [Cited by in F6Publishing: 124] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 2. | Colombo M, Sangiovanni A. Etiology, natural history and treatment of hepatocellular carcinoma. Antiviral Res. 2003;60:145-150. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 47] [Cited by in F6Publishing: 49] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 3. | Blum HE. Molecular therapy and prevention of hepatocellular carcinoma. Hepatobiliary Pancreat Dis Int. 2003;2:11-22. [PubMed] [Cited in This Article: ] |
| 4. | Galibert F, Mandart E, Fitoussi F, Tiollais P, Charnay P. Nucleotide sequence of the hepatitis B virus genome (subtype ayw) cloned in E. coli. Nature. 1979;281:646-650. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 723] [Cited by in F6Publishing: 769] [Article Influence: 17.1] [Reference Citation Analysis (0)] |
| 5. | Dong J, Cheng J. Study on definition of pre-pre-S region in hepatitis B virus genome. Shijie Huaren Xiaohua Zazhi. 2003;8:1091-1096. [Cited in This Article: ] |
| 6. | Yang Q, Dong J, Cheng J, Liu Y, Hong Y, Wang JJ, Zhuang SL. Definition of pre-pre-S promoter sequence from hepatttis B virus genome and identtfication of its transcription activity. Jiefangjun Yixue Zazhi. 2003;9:761-762. [Cited in This Article: ] |
| 7. | Liu Y, Cheng J, Lu YY, Li K. Studying of genes transactivated by hepatitis B viral protein. Shijie Huaren Xiaohua Zazhi. 2002;10:217-219. [Cited in This Article: ] |
| 8. | Maeng CY, Oh MS, Park IH, Hong HJ. Purification and structural analysis of the hepatitis B virus preS1 expressed from Escherichia coli. Biochem Biophys Res Commun. 2001;282:787-792. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 31] [Cited by in F6Publishing: 32] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 9. | Ji D, Cheng J, Wang JJ, Dong j, Guo J, Liu Y, Yang Q, Dang XY, Wang CH. Cloning of genes transactivated by pre-S1 protein of hepatitis B virus using suppression subtractive hybridization technique. Weichangbing He ganbingxue Zazhi. 2004;13:3-8. [Cited in This Article: ] |
| 10. | Borchani-Chabchoub I, Gargouri A, Mokdad-Gargouri R. Genotyping of Tunisian hepatitis B virus isolates based on the sequencing of preS2 and S regions. Microbes Infect. 2000;2:607-612. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 34] [Cited by in F6Publishing: 40] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 11. | Tai PC, Suk FM, Gerlich WH, Neurath AR, Shih C. Hypermodification and immune escape of an internally deleted middle-envelope (M) protein of frequent and predominant hepatitis B virus variants. Virology. 2002;292:44-58. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 78] [Cited by in F6Publishing: 84] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 12. | Diatchenko L, Lau YF, Campbell AP, Chenchik A, Moqadam F, Huang B, Lukyanov S, Lukyanov K, Gurskaya N, Sverdlov ED. Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc Natl Acad Sci USA. 1996;93:6025-6030. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2262] [Cited by in F6Publishing: 1997] [Article Influence: 71.3] [Reference Citation Analysis (0)] |
| 13. | Diatchenko L, Lukyanov S, Lau YF, Siebert PD. Suppression subtractive hybridization: a versatile method for identifying differentially expressed genes. Methods Enzymol. 1999;303:349-380. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 283] [Cited by in F6Publishing: 302] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 14. | Nakajima A, Kataoka K, Hong M, Sakaguchi M, Huh NH. BRPK, a novel protein kinase showing increased expression in mouse cancer cell lines with higher metastatic potential. Cancer Lett. 2003;201:195-201. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 44] [Cited by in F6Publishing: 44] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 15. | Redondo M, Villar E, Torres-Muñoz J, Tellez T, Morell M, Petito CK. Overexpression of clusterin in human breast carcinoma. Am J Pathol. 2000;157:393-399. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 184] [Cited by in F6Publishing: 193] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 16. | Miyake H, Hara S, Arakawa S, Kamidono S, Hara I. Over expression of clusterin is an independent prognostic factor for nonpapillary renal cell carcinoma. J Urol. 2002;167:703-706. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 48] [Cited by in F6Publishing: 50] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 17. | Miyake H, Gleave M, Kamidono S, Hara I. Overexpression of clusterin in transitional cell carcinoma of the bladder is related to disease progression and recurrence. Urology. 2002;59:150-154. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 82] [Cited by in F6Publishing: 88] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 18. | Xie MJ, Motoo Y, Su SB, Mouri H, Ohtsubo K, Matsubara F, Sawabu N. Expression of clusterin in human pancreatic cancer. Pancreas. 2002;25:234-238. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 64] [Cited by in F6Publishing: 67] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 19. | Chen X, Halberg RB, Ehrhardt WM, Torrealba J, Dove WF. Clusterin as a biomarker in murine and human intestinal neoplasia. Proc Natl Acad Sci USA. 2003;100:9530-9535. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 92] [Cited by in F6Publishing: 97] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 20. | Wellmann A, Thieblemont C, Pittaluga S, Sakai A, Jaffe ES, Siebert P, Raffeld M. Detection of differentially expressed genes in lymphomas using cDNA arrays: identification of clusterin as a new diagnostic marker for anaplastic large-cell lymphomas. Blood. 2000;96:398-404. [PubMed] [Cited in This Article: ] |
| 21. | Kang YK, Hong SW, Lee H, Kim WH. Overexpression of clusterin in human hepatocellular carcinoma. Hum Pathol. 2004;35:1340-1346. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 75] [Cited by in F6Publishing: 83] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 22. | Martínez-Hernández D, Arenas Barbero J, Jaqueti Aroca J, Pérez-Piqueras FJ, Santos Sancho J, Cosín Ochaitia J, Gómez de Terreros FJ. Adenosine deaminase, acquired immunodeficiency syndrome (AIDS), and hepatitis B infection. Clin Chem. 1992;38:162-163. [PubMed] [Cited in This Article: ] |
| 23. | Ghaffar F, Carrick K, Rogers BB, Margraf LR, Krisher K, Ramilo O. Disseminated infection with varicella-zoster virus vaccine strain presenting as hepatitis in a child with adenosine deaminase deficiency. Pediatr Infect Dis J. 2000;19:764-766. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 42] [Cited by in F6Publishing: 43] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 24. | Breen EC, Tang K. Calcyclin (S100A6) regulates pulmonary fibroblast proliferation, morphology, and cytoskeletal organization in vitro. J Cell Biochem. 2003;88:848-854. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 83] [Cited by in F6Publishing: 85] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 25. | Ferrari S, Calabretta B, deRiel JK, Battini R, Ghezzo F, Lauret E, Griffin C, Emanuel BS, Gurrieri F, Baserga R. Structural and functional analysis of a growth-regulated gene, the human calcyclin. J Biol Chem. 1987;262:8325-8332. [PubMed] [Cited in This Article: ] |
| 26. | Filipek A, Heizmann CW, Kuźnicki J. Calcyclin is a calcium and zinc binding protein. FEBS Lett. 1990;264:263-266. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 41] [Cited by in F6Publishing: 44] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 27. | Yao N, Coryell L, Zhang D, Georgescu RE, Finkelstein J, Coman MM, Hingorani MM, O'Donnell M. Replication factor C clamp loader subunit arrangement within the circular pentamer and its attachment points to proliferating cell nuclear antigen. J Biol Chem. 2003;278:50744-50753. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 45] [Cited by in F6Publishing: 46] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 28. | Griffith JD, Lindsey-Boltz LA, Sancar A. Structures of the human Rad17-replication factor C and checkpoint Rad 9-1-1 complexes visualized by glycerol spray/low voltage microscopy. J Biol Chem. 2002;277:15233-15236. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 89] [Cited by in F6Publishing: 94] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 29. | Fan J, Hooker CW, McManus DP, Brindley PJ. A new member of the transmembrane 4 superfamily (TM4SF) of proteins from schistosomes, expressed by larval and adult Schistosoma japonicum. Biochim Biophys Acta. 1997;1329:18-25. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 12] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 30. | Hashida H, Takabayashi A, Tokuhara T, Hattori N, Taki T, Hasegawa H, Satoh S, Kobayashi N, Yamaoka Y, Miyake M. Clinical significance of transmembrane 4 superfamily in colon cancer. Br J Cancer. 2003;89:158-167. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 94] [Cited by in F6Publishing: 100] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 31. | Yoshihama M, Nakao A, Nguyen HD, Kenmochi N. Analysis of ribosomal protein gene structures: implications for intron evolution. PLoS Genet. 2006;2:e25. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 32] [Article Influence: 1.8] [Reference Citation Analysis (0)] |