Copyright
©2006 Baishideng Publishing Group Co.
World J Gastroenterol. Aug 14, 2006; 12(30): 4836-4842
Published online Aug 14, 2006. doi: 10.3748/wjg.v12.i30.4836
Published online Aug 14, 2006. doi: 10.3748/wjg.v12.i30.4836
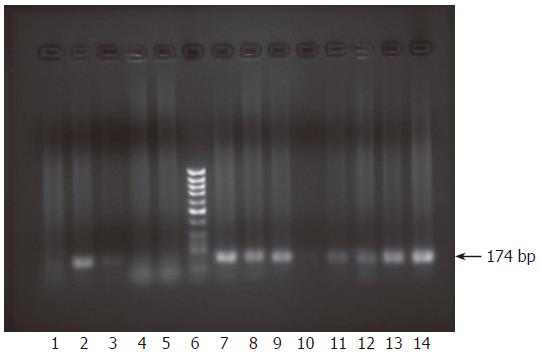
Figure 3 Monitoring infection of HepG2 cells using culture medium from primary infected cells by nested RT-PCR.
After incubation of HepG2 cells with infectious medium presumably containing exocytosed virions from primary infected cells, cells were carefully washed to get rid of any viral traces and the last wash was subjected to RNA extraction followed by nested RT-PCR. Results showed no amplification of positive strand products (lane 1). RNA was extracted from infected cells and supernatants at 3 d, 1, 2 and 4 wk post co-incubation with the infectious supernatant and subjected to nested RT-PCR to check for the presence of each viral strand. At 3 d the cells contained only the positive viral strand (lane 2) but the negative strand was undetectable (lane 3). However, the supernatants contained none of the strands, only primers used could be seen (lanes 4, 5). Lane 6 is molecular weight standard DNA marker (Ǿ-X-174/HaeIII; Q-BIOgene, Germany). At 1 wk post infection, both sense and antisense strands were detectable in the RNA extracted from the infected cells (lanes 7, 8), whereas the supernatant contained only the sense strand (lane 9) but not the antisense strand (lane 10). At 2 and 4 wk, RNA extracted from infected cells as well as the supernatants contained both positive and negative strands. Results of amplification for both positive and negative strands for cellular RNA are shown (2 wk, lanes 11, 12) and (4 wk, lanes 13, 14).
- Citation: El-Awady MK, Tabll AA, El-Abd YS, Bahgat MM, Shoeb HA, Youssef SS, Din NGBE, Redwan ERM, El-Demellawy M, Omran MH, El-Garf WT, Goueli SA. HepG2 cells support viral replication and gene expression of hepatitis C virus genotype 4 in vitro. World J Gastroenterol 2006; 12(30): 4836-4842
- URL: https://www.wjgnet.com/1007-9327/full/v12/i30/4836.htm
- DOI: https://dx.doi.org/10.3748/wjg.v12.i30.4836