Copyright
©2005 Baishideng Publishing Group Inc.
World J Gastroenterol. Dec 14, 2005; 11(46): 7277-7283
Published online Dec 14, 2005. doi: 10.3748/wjg.v11.i46.7277
Published online Dec 14, 2005. doi: 10.3748/wjg.v11.i46.7277
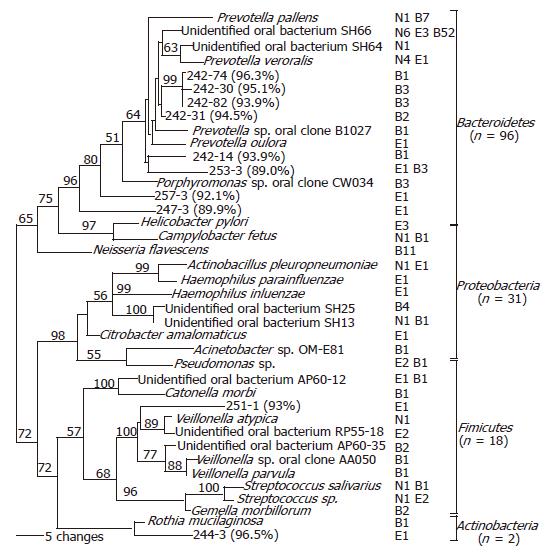
Figure 2 Phylogenetic analysis of bacterial 16S rDNA detected in the distal esophagus.
Sequences were aligned using ClustalW, and subjected to phylogenetic analysis using Paup 4.0b10 neighbor-joining analysis, based on HKY85 distance matrices. Bootstrap values (based on 500 replicates) are represented at each node when >50%, and the branch length index is represented below the phylogram. Names of bacterial species closest in homology with the detected 16S rDNA are located at the termination of each branch. “Unidentified” oral bacterial clones are potential bacterial species whose phylogenetic positions were designated by PCR-amplified 16S sequences only, rather than based on cultured organisms. The serial number of a clone followed by percentage of homology with the closest DNA sequence in GenBank is used as the species name for 16S rDNA sequence with <97% identity with all known DNA sequences. The frequency that a species was detected and its sources are shown following the species name. ‘N’ represents normal esophagus, ‘E’ represents esophagitis, and ‘B’ represents Barrett’s esophagus. The 39 species belong to four phyla, as shown at the right.
- Citation: Pei Z, Yang L, Peek RM, Levine JSM, Pride DT, Blaser MJ. Bacterial biota in reflux esophagitis and Barrett’s esophagus. World J Gastroenterol 2005; 11(46): 7277-7283
- URL: https://www.wjgnet.com/1007-9327/full/v11/i46/7277.htm
- DOI: https://dx.doi.org/10.3748/wjg.v11.i46.7277