Copyright
©2005 Baishideng Publishing Group Inc.
World J Gastroenterol. Jun 7, 2005; 11(21): 3255-3259
Published online Jun 7, 2005. doi: 10.3748/wjg.v11.i21.3255
Published online Jun 7, 2005. doi: 10.3748/wjg.v11.i21.3255
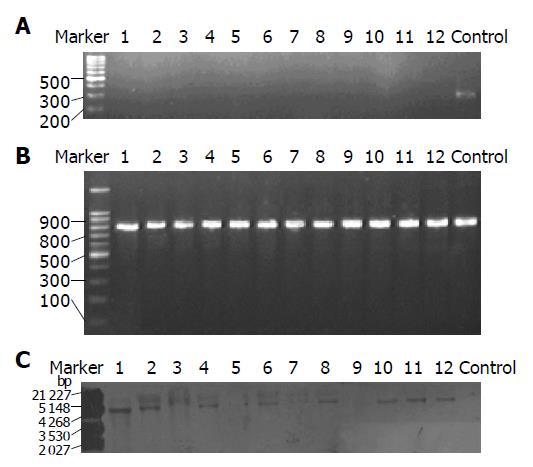
Figure 3 A: Result of PCR amplified for diagnosing homologous recombination strains of cagA gene.
Lanes 1-12 are 12 clones selected from chloramphenicol-resistant plates. Control is 26695 wild type. Twelve clones were all negative for diagnostic primers B1-2 while control is positive, suggesting that the flanking region of cagA gene of all the selected clones was deleted; B: Result of PCR amplified for positive control of homologous recombination strains of cagE gene. Lanes 1-12 are 12 clones selected from chloramphenicol-resistant plates. Control is 26695 wild type. Twelve clones were all positive of cagE gene; C: Result of Southern blot for diagnosing homologous recombination strains of cagA gene. Lanes 1-12 are 12 clones selected from chloramphenicol-resistant plates. Control is 26695 wild type. All the 12 clones except 3, 5, and 9 were positive hybridization with probes of chloramphenicol-resistant gene while control was negative.
-
Citation: Zeng X, He LH, Yin Y, Zhang MJ, Zhang JZ. Deletion of
cagA gene ofHelicobacter pylori by PCR products. World J Gastroenterol 2005; 11(21): 3255-3259 - URL: https://www.wjgnet.com/1007-9327/full/v11/i21/3255.htm
- DOI: https://dx.doi.org/10.3748/wjg.v11.i21.3255