Published online Apr 21, 2005. doi: 10.3748/wjg.v11.i15.2218
Revised: November 28, 2003
Accepted: February 18, 2004
Published online: April 21, 2005
AIM: To analyze the correlation between the protein expression of p16 and Rb genes in gastric carcinoma (GC), to investigate the role of p16 gene in invasion and lymph node metastasis of GC, and to examine the deletion and mutation in exon 2 of p16 gene in GC.
METHODS: The protein expression of p16 and Rb genes was examined by streptavidin-peroxidase conjugated method (S-P) in normal gastric mucosa, dysplastic gastric mucosa and GC. The deletion and mutation of p16 gene were examined by polymerase chain reaction (PCR) and polymerase chain reaction single strand conformation polymorphism (PCR-SSCP) respectively in normal gastric mucosa and GC.
RESULTS: The positive rates of P16 and Rb protein expression respectively were 96% (77/80) and 99% (79/80) in normal gastric mucosa, 92% (45/50) and 80% (40/50) in dysplastic gastric mucosa, 48% (58/122) and 60% (73/122) in GC. The positive rates of P16 and Rb protein expression in GC were significantly lower than that in normal gastric mucosa and dysplastic gastric mucosa (P<0.05). The positive rate of P16 protein expression in mucoid carcinoma (10%, 1/10) was significantly lower than that in poorly differentiated carcinoma (51%, 21/41), undifferentiated carcinoma (58%, 15/26) and signet ring cell carcinoma (62%, 10/16) (P<0.05). The positive rates of P16 protein in 30 cases of paired primary and lymph node metastatic GC were 47% (14/30) and 17% (5/30) respectively, being significantly lower in the later than in the former (P<0.05). There was no mutation in exon 2 of p16 gene in the 25 freshly resected primary GCs. But five cases in the 25 freshly resected primary GCs displayed deletion in exon 2 of p16 gene. The positive rate of both P16 and Rb proteins was 16% (14/90), and the negative rate of both P16 and Rb proteins was 8% (7/90) in 90 GCs. The rate of positive P16 protein with negative Rb protein was 33% (30/90). The rate of negative P16 protein with positive Rb protein was 43% (39/90). There was reverse correlation between P16 and Rb expression in 90 GCs (P<0.05).
CONCLUSION: The loss protein expression of p16 and Rb genes is related to GC. The loss expression of P16 protein is related to the histopathologic subtypes and lymph node metastasis of GC. Expression of P16 and Rb proteins in GC is reversely correlated. The deletion but not mutation in exon 2 of p16 gene may be involved in GC.
-
Citation: He XS, Rong YH, Su Q, Luo Q, He DM, Li YL, Chen Y. Expression of
p16 gene and Rb protein in gastric carcinoma and their clinicopathological significance. World J Gastroenterol 2005; 11(15): 2218-2223 - URL: https://www.wjgnet.com/1007-9327/full/v11/i15/2218.htm
- DOI: https://dx.doi.org/10.3748/wjg.v11.i15.2218
It is now widely accepted that carcinogenesis and progression of human Gastric carcinoma are related to the activation of proto-oncogenes and/or the inactivation of anti-oncogenes, and they are the results of genetic alteration accumulation. The p16 and Rb genes are tumor suppressor genes and participate in regulating the proliferation of normal cell negatively[1,2]. There were abnormal expressions of P16 and Rb proteins in a variety of cancer cell lines and primary tumors, such as osteosarcoma cell lines, renal cancer cell lines, lung cancer, brain tumor, esophageal cancer, breast cancer, hepatocellular carcinoma[3] and leukemogenesis[4]. In recent years, studies have revealed homozygous deletion and mutation of p16 gene, predominantly in exon 2, in various malignant tumors[5]. The frequency of p16 gene deletion and mutation is up to 75% in all kinds of human neoplasm, higher than that of the well-known p53 gene[1]. GC is one of the most common malignant tumors in China[6]. As with many other tumors, development of GC also involves multiple genes and stages, including some oncogenes and antioncogenes. However, till now, its carcinogenic molecular mechanism is not well clarified. In this paper, S-P immunohistochemical staining was used to detect the expression of P16 and Rb proteins in GC, dysplastic gastric mucosa and normal gastric mucosa. PCR and PCR-SSCP methods were used to detect deletion and mutation in exon 2 of p16 gene. We aimed at investigating the role of P16 protein in the carcinogenesis, progression, histologic types as well as biological behaviors of GC, to find a new marker for early diagnosis of GC, to discover the role of deletion and mutation in exon 2 of p16 gene in the carcinogenesis and progression of GC, and also to analyze the correlation of P16 and Rb proteins expression in GC.
All specimens were confirmed by pathological examination. Paraffin-embedded tissue specimens were collected from the pathology department and freshly resected specimens were collected from the surgery department of the First Affiliated Hospital of Nanhua University, among which there were 50 cases of dysplastic gastric mucosa, 80 cases of normal gastric mucosa (including 25 freshly resected specimens far away from cancer) and 122 GC (including 25 freshly resected specimens). In the 122 cases of GC, 29 were well-differentiated adenocarcinoma, 41 poorly-differentiated adenocarcinoma, 26 undifferentiated carcinoma, 16 signet ring cell carcinoma and 10 mucoid carcinoma. There were 81 men and 41 women, including 22 aged below 40 years, 69 aged from 41 to 59 years, and 31 older than 60 years (range 15-79, mean 56 years). Superficial muscles were invaded in 50 cases, deep muscles and the full layer in 72 cases. Sixty-nine cases had lymph node metastasis. The other 53 cases had no lymph node metastasis. Thirty cases of primary cancer and lymph node metastatic cancer respectively, were randomly selected and compared. According to Borrmann’s classification[7], 15 GCs were type I, 43 type II 47 type III and 17 type IV. The 25 freshly resected specimens, each containing cancer, cancer adjacent tissues and normal mucosa far away from cancer, were cut into 2-4 blocks under sterile conditions. Each block was 2-3 mm3 in size and stored at -70 °C for PCR and PCR-SSCP analyses. The rest of the tissues were fixed in 100 mL/L neutral formalin, resected, dehydrated, cleaned and paraffin-embedded. All paraffin-embedded tissues were cut into sequential slices at 5 μm thickness and mounted on to the glass slides that had been processed by poly-lys previously.
Rabbit-anti-human P16 protein polyclonal antibody, rabbit-anti-human Rb protein polyclonal antibody, streptavidin-peroxidase immunozator kit (S-P kit), and DAB were all purchased from Maxim Company, USA. Protease K (Merk, USA), SmaI, agarose gel, propylene acrylamide, N-N-sulmethyl bipropylene acrylamide, ammonium persulfate, xylene nitrile, bromophenol blue were purchased from Shanghai Sangon Company. PCR primers were synthesized by Shanghai Sangon. Primer sequences in exon 2 of p16 gene: sense 5’-TCTGACCATTCTGTTCTCTC-3’, antisense 5’-CTCAGCTTTGGAAGCTCTCA-3’, were used for PCR and PCR-SSCP. The fragment length of amplification was 384 bp. Primer sequences of β-actin served as an internal control for PCR: sense 5’-GCGGGGCGCCCCAGGCACCA-3’, antisense 5’-CTCCTTAATGTCACGCACGATTTC-3’. The fragment length of amplification was 548 bp. Experimental instruments included ultra low refrigerator of -70 °C (Japan), rotary sector (Germany), microscope (Japan), type 480 DNA amplificatory (PE, USA), type 901 ultraviolet spectrophotometer (PE, USA), type DYB vertical electrophoresis and various kinds of centrifuges (Liuyi, Beijing).
S-P immunohistochemical staining According to the specification of S-P kit: paraffin-embedded tissue slices were deparaffinized and hydrated for 20 min, endogenous peroxidase was blocked for 10 min, first antibody was added (rabbit-anti-human P16 protein polyclonal antibody or rabbit-anti-human Rb protein polyclonal antibody) for 8 h at 4 °C, bridge antibody was added for 15 min, enzyme labeled S-P reagents were added for 10 min, and was colored with DAB for 5 min, nucleolus was stained with hematoxylin for 3 min, dehydrated for 15 min, cleaned for 10 min, later was covered and observed under a microscope.
Genomic DNA extraction[8] Frozen tissue of 0.5 g was put into liquid nitrogen and powdered immediately, followed by addition of 10×buffer (10 mmol/L Tris-HCl, pH 8.0, 0.1 mol/L EDTA, pH 8.0, 5 g/L SDS) in 37 °C water for 1 h. At the same time, protease K was added to the mixture at a final concentration of 100 mg/L in 50 °C water for 3 h. After the mixtures were dissolved completely, the mixtures were reacted with 20 mg/L Rnase in 37 °C water for 1 h. It was mixed with saturated phony and bugged slightly for 10 min, then centrifuged and supernatant was extracted, and transferred to a cleaned plastic tube. The above processes were repeated thrice. The supernatant was added with 1/10 volume 3 mol/L NaAc and 2-2.5 times cold ethyl, and centrifuged. DNA was precipitated, ethyl removed. DNA was washed by 700 mL/L ethyl and centrifuged thrice, dried, resolved with TE and stored at -20 °C for use.
PCR amplification PCR was performed according to the reference[8] in 50 μL reaction mixture containing 0.1 μg of DNA template, 200 μmol/L each of dCTP, dATP, dGTP, dTTP, 0.25 μmol/L primer, PCR buffer (10 mmol/L Tris-HCl, pH 8.3, 1.5 μmol/L MgCl2, 50 mmol/L KCl, 100 mg/L gelatin). Then the reaction mixture was pre-denatured at 95 °C for 5 min and added with 1.5 μL of Taq DNA polymerase and 75 μL of mineral oil. These samples were subjected to 30 amplification cycles, each consisting of denaturation at 95 °C for 1 min, primer annealing at 60 °C for 1 min, and extension at 72 °C for 1 min. Finally, the samples were subjected to a further extension at 72 °C for 5 min. A 5 μL of PCR product was electrophoresed on agarose gel (20 g/L), then observed and photographed under ultraviolet light. No products of PCR amplification suggested loss of homozygosis of p16 gene.
PCR-SSCP analysis[8] A 5 μL of PCR product digested by SmaI was mixed with 5 μL of denatured dissolution (950 mL/L formamide, 20 mmol/L EDTA, 0.05% bromophenol blue, 0.5 g/L xylene nitrile) and denatured at 95 °C for 5 min, and then cooled on ice. Solution processed as above was added to the gel containing 80 g/L polypropylene acrylamide, vertically electrophoresed at 100 V for 4 h. The gel stained with silver was fixed in 100 mL/L alcohol for 10 min, oxidized in 100 g/L nitric acid for 3 min, drip washed for 1 min with double distilled water, stained in 12 mmol/L silver nitric acid for 20 min, again drip washed for 1 min with double distilled water, showed appropriately colored in 0.028 moL anhydrous sodium carbonate and 0.19 mL/L formalin, reduction response was ended by 100 mL/L glacial acetic acid, drip washed with double distilled water. The results were analyzed and photographed. The abnormal traces found in PCR-SSCP were considered gene mutation.
Immunohistochemical determination The brown-yellow staining of nucleus or nucleus and cytoplasm was considered positive; (-) indicates no positively stained cell or only plasma stained or the number of nuclear stained positive cells less than 1; (+) indicates the cells stained weakly or the number of stained cells less than 25%; (++) indicates the cells stained moderately or the stained cells about 26-50%; (+++) indicates cells stained strongly or the number of stained cells more than 50%. Positive nuclear staining in more than two cells (under high power microscope) was considered to be positive. No folding, and edging-effect fields were chosen during calculation of 100 cells per five fields. The assessment was performed by two observers. P16 protein expression in positively confirmed cervical carcinoma served as positive control. The first antibody was replaced with PBS for negative control.
χ2 test was used to analyze the data. P value less than 0.05 was considered statistically significant.
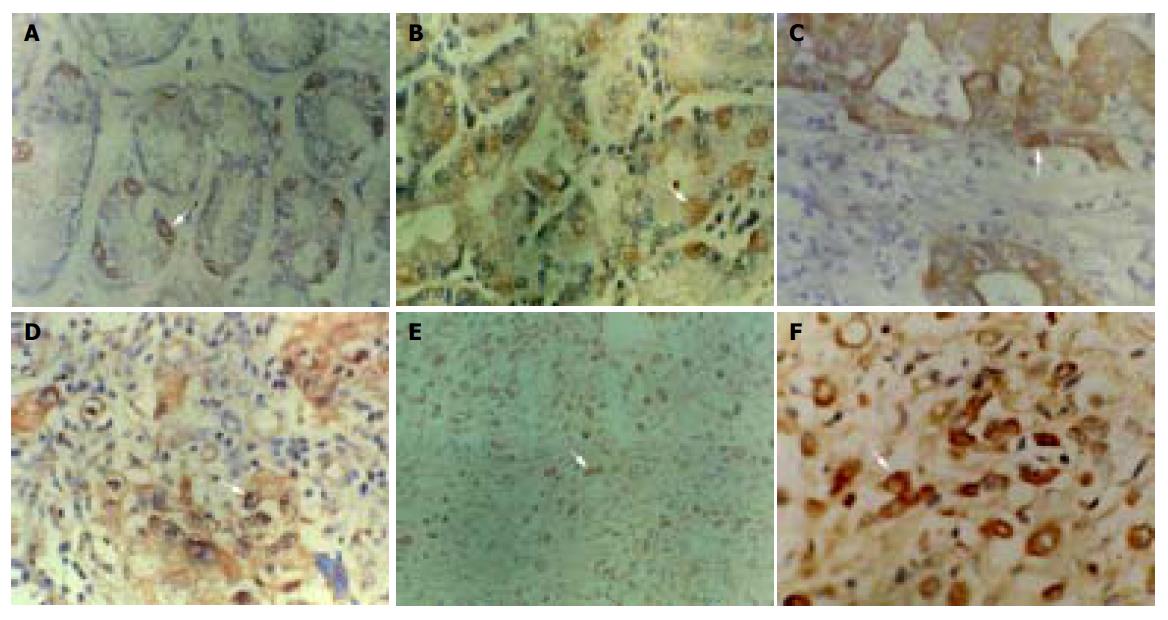
P16 protein expression could only be seen in particular adenoepithelial cells. We did not find positive staining in mucosal epithelial cells, matrix fibrocytes, lymphocytes and smooth myocytes. The positive rate of P16 protein expression was 96% (77/80) in normal gastric mucosa (Figure 1A) and 90% (45/50) in dysplastic gastric mucosa (Figure 1B). There was no significant difference in these mucosa. But in GC, the positive rate of P16 protein expression was 48% (58/122) (Figures 1C-1F), lower than that in normal and dysplastic mucosa (P<0.05, Table 1). The positive rate of P16 protein expression was 38%, 51%, 58%, 62% and 10% in well-differentiated adenocarcinoma, poorly-differentiated adenocarcinoma, undifferentiated carcinoma, signet ring cell carcinoma and mucoid carcinoma, respectively. The P16 protein expression in mucoid carcinoma was significantly lower than that in signet ring cell carcinoma, undifferentiated carcinoma and poorly-differentiated adenocarcinoma (P<0.05, Table 2). The positive rate of P16 protein expression was 48% (24/50) in GC with superficial muscle layer invasion and 47% (34/72) in GC with deep muscle and full layer invasion. There was no apparent relevance between P16 protein expression and the depth of invasion (Table 2). In 30 cases of paired primary and lymph node metastatic GC, the rate of P16 protein expression in lymph node metastatic cancer (17%, 5/30) was significantly lower than that of primary cancer (47%, 14/30) (P<0.05, Table 3).
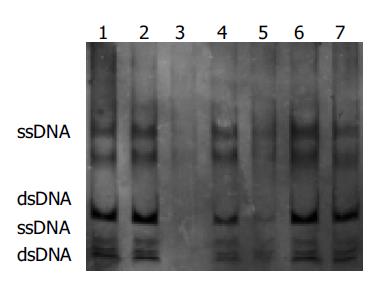
Among 25 freshly resected GCs, there were seven well-differentiated adenocarcinoma, 13 poorly-differentiated adenocarcinoma, three undifferentiated carcinoma, one signet ring cell carcinoma and one mucoid carcinoma. Cancer adjacent tissue and normal gastric mucosa were taken at the same time. The PCR amplification showed no product in one case of well-differentiated adenocarcinoma, one case of poorly-differentiated adenocarcinoma and one case of mucoid carcinoma. Few products found in one case of well-differentiated adenocarcinoma and one case of poorly-differentiated adenocarcinoma. But the remaining 20 cases of GC, cancer adjacent gastric mucosa and normal gastric mucosa showed products of PCR amplification. All experiments were repeated thrice. The results were identical. No product of PCR amplification could indicate the loss of homozygosis of p16 gene, few products of PCR amplification showed possible loss of heterozygosis or homozygosis of p16 gene contaminated with normal mucosa (Figure 2). Four of these five cases showed negative expression for P16 protein and one case showed weak expression detected with immunohistochemical staining. No gene mutation was observed by PCR-SSCP analysis after the PCR amplification products were cut with SmaI (Figure 3 and Table 1).
The positive rate of Rb protein expression in normal gastric mucosa was 99% (79/80), 80% (40/50) in dysplastic gastric mucosa, and 60% (73/122) in GC (Figure 4). The positive rate of Rb protein expression in GC was significantly lower than that in normal gastric mucosa and dysplastic mucosa (P<0.05, Table 4). There was no significant difference between the normal gastric mucosa and dysplastic gastric mucosa (P>0.05, Table 4).
Ninety paired GCs were selected from 122 cases of GC to make a comparison analysis. The positive expression of both P16 and Rb proteins was 16% (14/90), the negative expression of both P16 and Rb proteins was 8% (7/90). The positive expression of P16 with negative expression of Rb was 33% (30/90). The negative expression of P16 with positive expression of Rb was 43% (39/90). The results suggested negative correlation between P16 and Rb proteins expression in GC (P<0.05, Table 5).
The development of human cancers including GC is a multi-step process and phenotypic changes during cancer progression reflect the sequential accumulation of genetic alterations in cells[9]. Both p16 and Rb genes are tumor suppressor genes. They play important roles in the regulation of the cell cycle. The proteins of these two genes, P16 and pRb respectively, inhibit cell progression from G1 to S phase[1,2]. Dephosphorylation retinoblastoma protein (Rb) inactivates the transcription factors such as E2F1, an important factor for the transition from G1 to S phase, thereby arrests cells in G0/G1 phase, resulting in suppressed cell division and proliferation. When Rb protein is phosphorylated, several transcription factors are released, which induce the cell from G1 to S phase rapidly, resulting in excessive proliferation of cell. P16 has been shown to exert its function through inhibition of cyclin-dependent kinase 4(CDK4) mediated phosphorylation of pRb. Functional loss of p16 might result in nonregulation of CDK4 activity, leading to persistent pRb phosphorylation and uncontrolled cellular proliferation[1,2]. Abnormalities of p16 and Rb genes are frequent molecular events in human cancer[10-13]. Some investigations demonstrated increased cell cycle arrest, growth inhibition and apoptosis after adenovirus-mediated transduction of p16 gene in gliomas, lung, pancreas, liver, head and neck tumor cell lines[1,2]. All these findings indicate that p16 and Rb are important tumor suppressor genes.
In human GC, loss of p16 expression is common. Our result showed that the positive rate of P16 protein expression in GC was remarkably lower than that in dysplastic gastric mucosa and normal gastric mucosa. This is in accordance with observations by others[1-3]. These results indicated that P16 loss expression was characteristically associated with tumor progression in GC. Several other studies have shown that re-expression of p16 in various tumor cell lines was sufficient to cause arrest of the cells in G1 phase[14,15]. In our study, there was no significant difference between the normal mucosa and the dysplastic mucosa of stomach in P16 protein expression. Interestingly, the expression quantity of P16 protein increased from normal mucosa to precancerous lesions and GC. Nguyen et al[16], found that the level of p16 mRNA in part of prostate carcinoma was higher than that in prostate tissues. They also showed that P16 protein expression increased with the progression of the pathologic lesion. This change might inhibit cell proliferation. Our results showed that the positive rate of P16 protein expression was significantly lower in mucoid carcinoma than that in poorly-differentiated adenocarcinoma, undifferentiated carcinoma and signet cell carcinoma. It suggested that the alteration of p16 gene was different between various histologic types of GC. The discrepancy of P16 protein expression also existed between various histologic types of lung cancer, esophageal cancer and gliomas[10,17]. In our investigation, P16 protein expression was not significantly related to sex, age, the depth of invasion and Borrmann’s classification. However, decreased expression of p16 was found more frequently in lymph node metasatic GC than in primary GC. It was demonstrated that the loss of expression of P16 protein contributed to tumor progression with lymph node metastasis, which is in agreement with previous reports[6]. Some studies indicated that the loss of p16 expression was associated with aggressive phenotype and poor prognosis of several tumors[18]. The positive expression of P16 protein could merely be observed in partial adeno-epithelial cells of normal and dysplastic gastric mucosa, and were weakly positive or undetectable in gastric mucosa epithelium cells, interstitial lymphocytes, fibroblasts and smooth muscle cells, which were contrary to some published reports[19]. Nevertheless, P16 protein expression was undetectable in neoplasmic stroma[20], normal lung tissue[21] and normal urethral epithelium cells[19]. This might attribute to a paucity of P16 molecule in G0/G1 phase cells[22] or a short half-life of P16 protein[23].
Some studies have shown that deletion and mutation of p16 gene are important mechanisms responsible for the dysfunction of tumor suppressor genes[24]. In our present study, deletion in exon 2 of p16 gene was detected in five GC tissues. But PCR amplification products appeared in the rest of the 20 cases of GC, normal gastric mucosa and cancer adjacent gastric mucosa, suggesting an association between deletion of p16 gene and GC. Previous studies have shown that deletion of p16 gene is associated with the degree of differentiation and metastasis of GC. Our observation failed to demonstrate the similar trend. It was likely that only exon 2 was examined due to inadequate specimens or other unknown factors. The mutation of p16 gene was not found by SSCP analysis of digestion product of PCR amplification. The results revealed that point mutation of p16 gene was rare during gastric carcinogenesis. It coincides with previous findings[25,26]. It was also implied that the frequency of p16 gene deletion was lower than that of loss of P16 protein expression. Some other uncertain mechanisms might exist in the regulation of p16 gene expression. Many studies suggested that hypermethylation of p16 gene was the major process for its inactivation in GC and an important mechanism in gastric carcinogenesis[27]. Further studies are needed to prove this.
Immunohistochemical analysis showed that the positive rate of pRb was significantly lower in GC than in normal gastric mucosa and dysplastic gastric mucosa. The result indicated GC carcinogenesis was probably related with the loss of pRb expression, which was consistent with other reports[28]. Serrano et al[1], have proposed that physiological inactivation of Rb during G1 phase leads to increased p16 expression in order to limit CDK4 activity. Genetic inactivation of Rb would also stimulate cell to increase p16 expression in an ultimately unnecessary attempt to inhibit CDK4. This negative feedback model predicts that Rb-negative tumors would have high levels of p16, while Rb-positive tumors might require decreased amounts of functional p16 in order to achieve a level of CDK4 activity sufficient for Rb inactivation. Shapiro et al[21], also confirmed the inverse reciprocity between Rb inactivation and p16 expression in lung cancer. We therefore sought to determine whether there was a specific correlation between p16 gene and Rb gene in GC. We analyzed the relationship between P16 and pRb expression in GC. The result indicated that there was not only loss of P16 and Rb proteins expression, but also negative correlation between P16 and Rb expression in human GC which is consistent with others[29]. These support the hypothesis that p16 and Rb genes adjust with each other by negative feedback in cell cycle regulation and indicate that the alteration of p16 and Rb gene expression may be involved in carcinogenesis and progression of human GC.
| 1. | Serrano M, Hannon GJ, Beach D. A new regulatory motif in cell-cycle control causing specific inhibition of cyclin D/CDK4. Nature. 1993;366:704-707. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2515] [Cited by in RCA: 2555] [Article Influence: 79.8] [Reference Citation Analysis (0)] |
| 2. | Kamb A, Gruis NA, Weaver-Feldhaus J, Liu Q, Harshman K, Tavtigian SV, Stockert E, Day RS, Johnson BE, Skolnick MH. A cell cycle regulator potentially involved in genesis of many tumor types. Science. 1994;264:436-440. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1955] [Cited by in RCA: 1887] [Article Influence: 60.9] [Reference Citation Analysis (0)] |
| 3. | Peng CY, Chen TC, Hung SP, Chen MF, Yeh CT, Tsai SL, Chu CM, Liaw YF. Genetic alterations of INK4alpha/ARF locus and p53 in human hepatocellular carcinoma. Anticancer Res. 2002;22:1265-1271. [PubMed] |
| 4. | Mitani K. Molecular mechanisms in leukemogenesis. Gan To Kagaku Ryoho. 2002;29:1107-1112. [PubMed] |
| 5. | Sasaki S, Kitagawa Y, Sekido Y, Minna JD, Kuwano H, Yokota J, Kohno T. Molecular processes of chromosome 9p21 deletions in human cancers. Oncogene. 2003;22:3792-3798. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 59] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 6. | He XS, Su Q, Chen ZC, He XT, Che SY. Expression, Mutation and Deletion of p16 Gene in gastric carcinoma. Aizheng. 2001;20:468-473. |
| 7. | Borrman R. Geschwulste des Magens und Duodenums. In Henke F. Lubarsch O. eds: Handbuch der Speziellen Pathologischen Anatomie und Histologie. Berlin Julius Springer 1926; 864-871. |
| 8. | Sambrook J, Frisch EF, Maniatis T. Molecular Cloning, A Laboratory Manual 2nded, Cold Spring Harbor Laboratory Press 1989. . |
| 9. | Yasui W, Yokozaki H, Fujimoto J, Naka K, Kuniyasu H, Tahara E. Genetic and epigenetic alterations in multistep carcinogenesis of the stomach. J Gastroenterol. 2000;35 Suppl 12:111-115. [PubMed] |
| 10. | Kratzke RA, Greatens TM, Rubins JB, Maddaus MA, Niewoehner DE, Niehans GA, Geradts J. Rb and p16INK4a expression in resected non-small cell lung tumors. Cancer Res. 1996;56:3415-3420. [PubMed] |
| 11. | Jarrard DF, Modder J, Fadden P, Fu V, Sebree L, Heisey D, Schwarze SR, Friedl A. Alterations in the p16/pRb cell cycle checkpoint occur commonly in primary and metastatic human prostate cancer. Cancer Lett. 2002;185:191-199. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 44] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 12. | Mobley SR, Liu TJ, Hudson JM, Clayman GL. In vitro growth suppression by adenoviral transduction of p21 and p16 in squamous cell carcinoma of the head and neck: a research model for combination gene therapy. Arch Otolaryngol Head Neck Surg. 1998;124:88-92. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 28] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 13. | Tsujie M, Yamamoto H, Tomita N, Sugita Y, Ohue M, Sakita I, Tamaki Y, Sekimoto M, Doki Y, Inoue M. Expression of tumor suppressor gene p16(INK4) products in primary gastric cancer. Oncology. 2000;58:126-136. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 36] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 14. | Arap W, Nishikawa R, Furnari FB, Cavenee WK, Huang HJ. Replacement of the p16/CDKN2 gene suppresses human glioma cell growth. Cancer Res. 1995;55:1351-1354. [PubMed] |
| 15. | Frizelle SP, Grim J, Zhou J, Gupta P, Curiel DT, Geradts J, Kratzke RA. Re-expression of p16INK4a in mesothelioma cells results in cell cycle arrest, cell death, tumor suppression and tumor regression. Oncogene. 1998;16:3087-3095. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 84] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 16. | Nguyen TT, Nguyen CT, Gonzales FA, Nichols PW, Yu MC, Jones PA. Analysis of cyclin-dependent kinase inhibitor expression and methylation patterns in human prostate cancers. Prostate. 2000;43:233-242. [PubMed] [DOI] [Full Text] |
| 17. | Nishikawa R, Furnari FB, Lin H, Arap W, Berger MS, Cavenee WK, Su Huang HJ. Loss of P16INK4 expression is frequent in high grade gliomas. Cancer Res. 1995;55:1941-1945. [PubMed] |
| 18. | Kawabuchi B, Moriyama S, Hironaka M, Fujii T, Koike M, Moriyama H, Nishimura Y, Mizuno S, Fukayama M. p16 inactivation in small-sized lung adenocarcinoma: its association with poor prognosis. Int J Cancer. 1999;84:49-53. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 19. | Reznikoff CA, Yeager TR, Belair CD, Savelieva E, Puthenveettil JA, Stadler WM. Elevated p16 at senescence and loss of p16 at immortalization in human papillomavirus 16 E6, but not E7, transformed human uroepithelial cells. Cancer Res. 1996;56:2886-2890. [PubMed] |
| 20. | Geradts J, Hruban RH, Schutte M, Kern SE, Maynard R. Immunohistochemical p16INK4a analysis of archival tumors with deletion, hypermethylation, or mutation of the CDKN2/MTS1 gene. A comparison of four commercial antibodies. Appl Immunohistochem Mol Morphol. 2000;8:71-79. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 25] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 21. | Shapiro GI, Edwards CD, Kobzik L, Godleski J, Richards W, Sugarbaker DJ, Rollins BJ. Reciprocal Rb inactivation and p16INK4 expression in primary lung cancers and cell lines. Cancer Res. 1995;55:505-509. [PubMed] |
| 22. | Tam SW, Shay JW, Pagano M. Differential expression and cell cycle regulation of the cyclin-dependent kinase 4 inhibitor p16Ink4. Cancer Res. 1994;54:5816-5820. [PubMed] |
| 23. | Shapiro GI, Park JE, Edwards CD, Mao L, Merlo A, Sidransky D, Ewen ME, Rollins BJ. Multiple mechanisms of p16INK4A inactivation in non-small cell lung cancer cell lines. Cancer Res. 1995;55:6200-6209. [PubMed] |
| 24. | Giroux MA, Audrezet MP, Metges JP, Lozac'h P, Volant A, Nousbaum JB, Labat JP, Gouérou H, Ferec C, Robaszkiewicz M. Infrequent p16/CDKN2 alterations in squamous cell carcinoma of the oesophagus. Eur J Gastroenterol Hepatol. 2002;14:15-18. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 25. | Günther T, Schneider-Stock R, Pross M, Manger T, Malfertheiner P, Lippert H, Roessner A. Alterations of the p16/MTS1-tumor suppressor gene in gastric cancer. Pathol Res Pract. 1998;194:809-813. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 17] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 26. | Sakata K, Tamura G, Maesawa C, Suzuki Y, Terashima M, Satoh K, Eda Y, Suzuki A, Sekiyama S, Satodate R. Loss of heterozygosity on the short arm of chromosome 9 without p16 gene mutation in gastric carcinomas. Jpn J Cancer Res. 1995;86:333-335. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 21] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 27. | Ficorella C, Cannita K, Ricevuto E, Toniato E, Fusco C, Sinopoli NT, De Galitiis F, Di Rocco ZC, Porzio G, Frati L. P16 hypermethylation contributes to the characterization of gene inactivation profiles in primary gastric cancer. Oncol Rep. 2003;10:169-173. [PubMed] |
| 28. | Ogawa M, Maeda K, Chung YS, Onoda N, Kato Y, Nakata B, Sowa M. Correlation between expression of RB protein and prognosis of gastric cancer. Gan To Kagaku Ryoho. 1996;23 Suppl 2:148-150. [PubMed] |
| 29. | Lee WA, Woo DK, Kim YI, Kim WH. p53, p16 and RB expression in adenosquamous and squamous cell carcinomas of the stomach. Pathol Res Pract. 1999;195:747-752. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 21] [Article Influence: 0.8] [Reference Citation Analysis (0)] |