Copyright
©The Author(s) 2004.
World J Gastroenterol. Dec 15, 2004; 10(24): 3553-3558
Published online Dec 15, 2004. doi: 10.3748/wjg.v10.i24.3553
Published online Dec 15, 2004. doi: 10.3748/wjg.v10.i24.3553
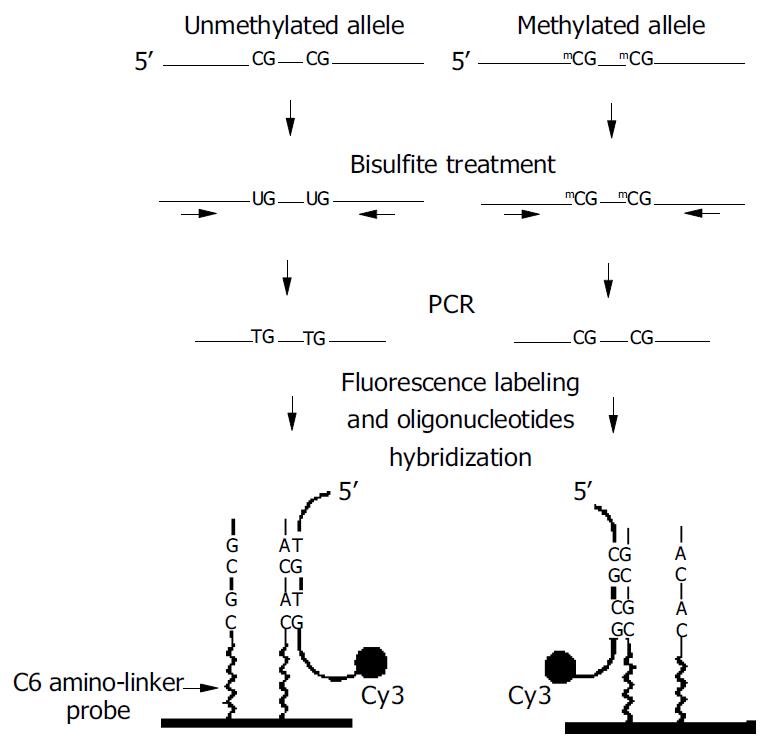
Figure 1 Schematic outline for analysis of DNA methylation based on oligonucleotide microarray[20].
Genomic DNA was bisulfite treated and amplified by PCR for a specific CpG island region of interest. The amplified product was labeled with Cy3 fluorescence dye and hybridized to oligonucleotide probes at-tached to a glass surface. At left an oligonucleotide probe was designed to form a perfect match with a target DNA containing the unmethylated allele. At right a probe was designed to form a perfect match with the methylated DNA target.
- Citation: Hou P, Shen JY, Ji MJ, He NY, Lu ZH. Microarray-based method for detecting methylation changes of p16Ink4a gene 5’-CpG islands in gastric carcinomas. World J Gastroenterol 2004; 10(24): 3553-3558
- URL: https://www.wjgnet.com/1007-9327/full/v10/i24/3553.htm
- DOI: https://dx.doi.org/10.3748/wjg.v10.i24.3553