Copyright
©The Author(s) 2004.
World J Gastroenterol. Dec 1, 2004; 10(23): 3441-3454
Published online Dec 1, 2004. doi: 10.3748/wjg.v10.i23.3441
Published online Dec 1, 2004. doi: 10.3748/wjg.v10.i23.3441
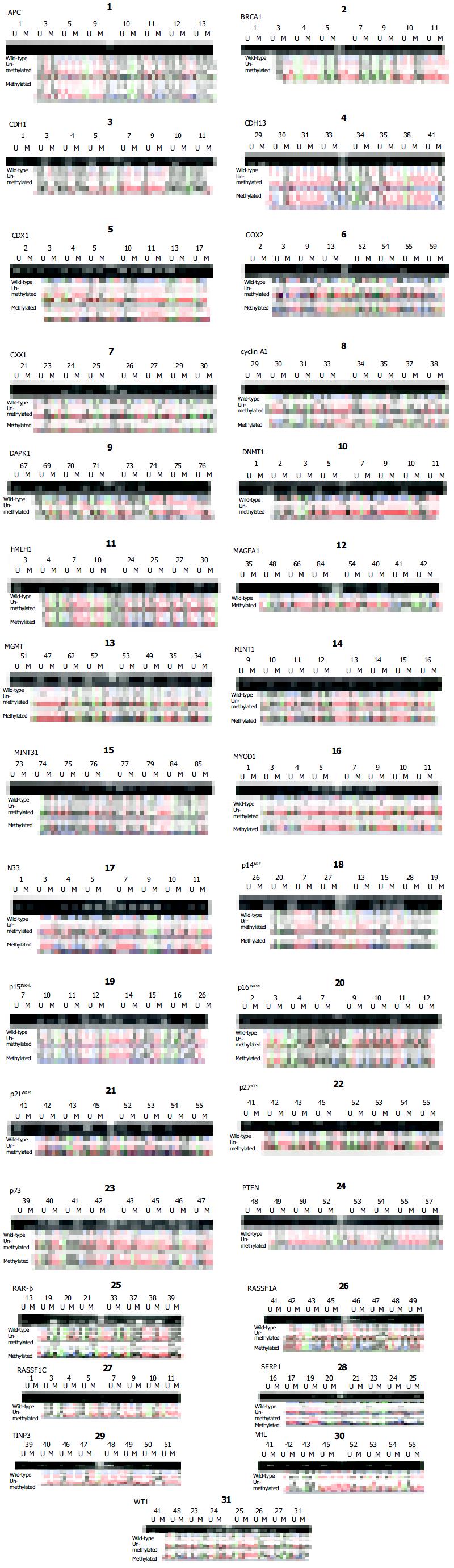
Figure 1 Methylation profiles of the promoter CpG islands of 31 genes in CRC.
Both electrophoretic patterns of the representa-tive PCR products of each of 31 targets (indicated respectively, at the top of figures) and the sequencing verification of the one representative PCR product are presented. To indicate the methylation status, the sequenced data are aligned with the wild-type sequence. 1Size markers, the bands of 250 bp and 100 bp are shown. U, the unmethylated; M, hypermethylated. Panels: 1, APC, 2, BRCA1, 3, CDH1, 4, CDH13, 5, CDX1, 6, COX2, 7,CXX1, 8, cyclin A1, 9, DPAK1, 10, DNMT1, 11, hMLH1, 12, MAGEA1, 13, MGMT, 14, MINT1, 15, MINT31, 16, MYOD1, 17, N33, 18, p14ARF, 19, p15INK4b, 20, p16INK4a 21, p21WAF1, 22, p27KIP1, 23, p73, 24, PTEN, 25, RAR-b, 26, RASSF1A, 27, RASSF1C, 28, SFRP1, 29, TIMP3, 30, VHL, and 31, WT1.
- Citation: Xu XL, Yu J, Zhang HY, Sun MH, Gu J, Du X, Shi DR, Wang P, Yang ZH, Zhu JD. Methylation profile of the promoter CpG islands of 31 genes that may contribute to colorectal carcinogenesis. World J Gastroenterol 2004; 10(23): 3441-3454
- URL: https://www.wjgnet.com/1007-9327/full/v10/i23/3441.htm
- DOI: https://dx.doi.org/10.3748/wjg.v10.i23.3441