Copyright
©The Author(s) 2023.
World J Gastroenterol. Aug 14, 2023; 29(30): 4642-4656
Published online Aug 14, 2023. doi: 10.3748/wjg.v29.i30.4642
Published online Aug 14, 2023. doi: 10.3748/wjg.v29.i30.4642
Figure 1 Identification of 116 upregulated tRNA-derived fragments and 95 downregulated tRNA-derived fragments in acute pancreatitis by the differential expression analysis.
A: Heatmap of the differentially expressed tRNA-derived fragments (DE-tRFs); B: Volcano plot for the 116 upregulated and 95 downregulated DE-tRFs; C: Gene expression of the five most significant DE-tRFs using reverse transcription quantitative polymerase chain reaction. Error bars represent the mean ± SD, a and b represent different expression levels, the same letter represents no significant difference between groups (P > 0.05), different letters represent significant difference between groups (P < 0.05). AP: Acute pancreatitis; tRF: tRNA-derived fragment.
Figure 2 Effects of tRNA-derived fragments 36 on acute pancreatitis progression in MCP-83 cells.
A: Gene expression of tRNA-derived fragments 36 using reverse transcription quantitative polymerase chain reaction; B-D: The expression levels of inflammatory factors in the acute pancreatitis (AP) cell model, including interleukin-1β (B), interleukin-6 (C), and tumor necrosis factor-α (D); E: The expression levels of lipase in the AP cell model; F: The expression levels of amylase in the AP cell model; G: Cell viability examination using Cell Counting Kit-8 assays; H: Cell death examination using terminal deoxynucleotidyl transferase dUTP nick-end labeling analysis. Error bars represent the mean ± SD. a, b, and c represent different expression levels, the same letter represents no significant difference between groups (P > 0.05), different letters represent significant difference between groups (P < 0.05). tRF-36: tRNA-derived fragments 36; IL: Interleukin; TNF: Tumor necrosis factor; AMS: Amylase; NC: Normal control; DAPI: 4’,6-diamidino-2-phenylindole.
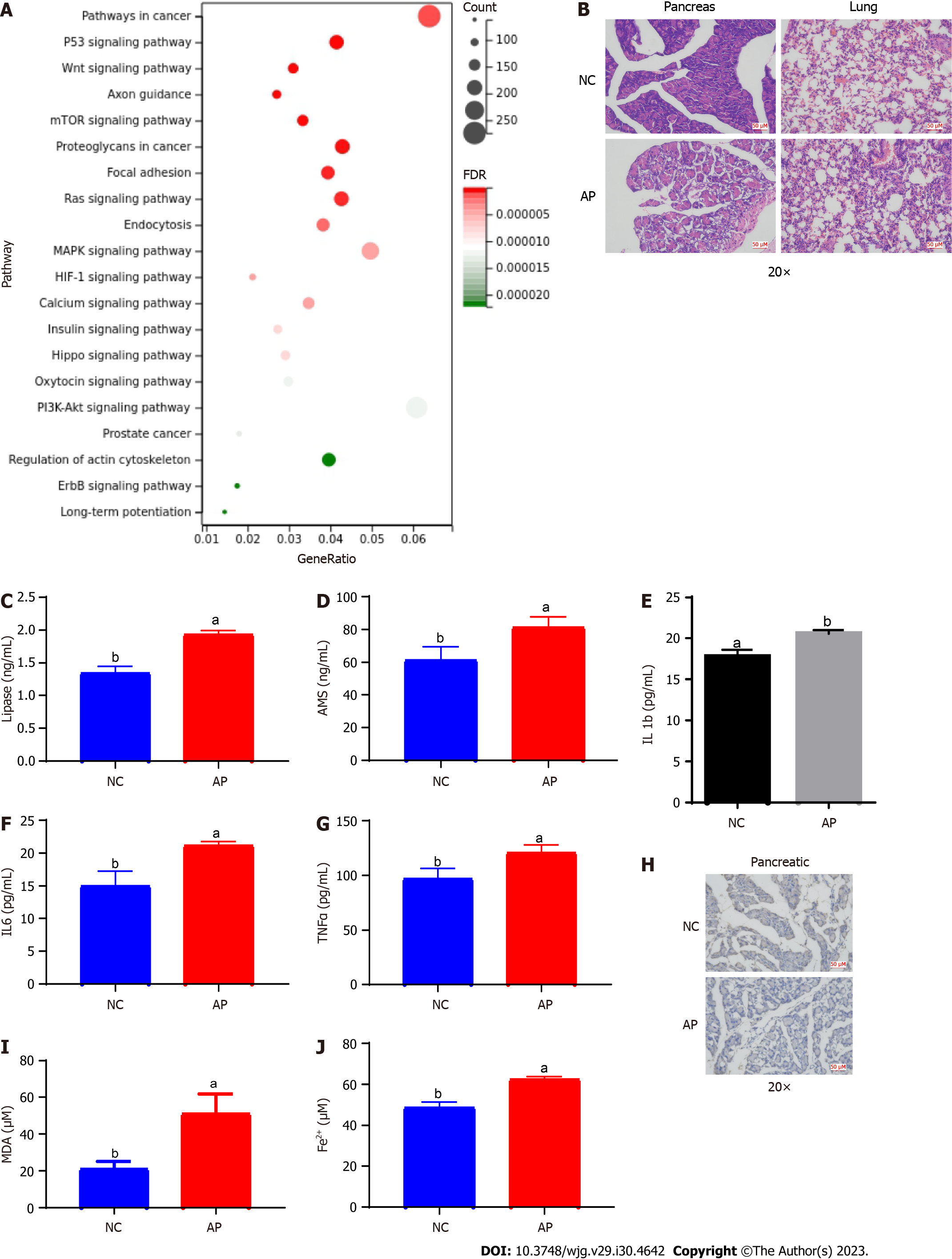
Figure 3 Evaluation of the target genes of tRNA-derived fragments 36 for the cell ferroptosis process in acute pancreatitis.
A: Functional enrichment analysis for the target genes of tRNA-derived fragments 36; B: Hematoxylin and eosin staining results of lung and pancreatic tissues in the acute pancreatitis (AP) mouse model; C and D: The serum levels of lipase and amylase in the AP mouse model; E-G: The serum levels of the inflammatory factors interleukin-1β (E), interleukin-6 (F), and tumor necrosis factor-α (G); H: The expression of ferritin in the mouse pancreatic tissues with AP; I: The levels of malondialdehyde in mouse pancreatic tissues with AP; J: The ferric ion concentration in mouse pancreatic tissues with AP. a and b represent different expression levels, the same letter represents no significant difference between groups (P > 0.05), different letters represent significant difference between groups (P < 0.05). tRF-36: tRNA-derived fragments 36; IL: Interleukin; TNF: Tumor necrosis factor; NC: Normal control; mTOR: Mechanistic target of rapamycin; HIF-1: Hypoxia-inducible factor 1; FDR: False discovery rate; PI3K-AKT: Phosphatidylinositol-4,5-bisphosphate 3-kinase-protein kinase B; ErbB: Erythroblastic oncogene B; AP: Acute pancreatitis; MDA: Malondialdehyde; AMS: Amylase.
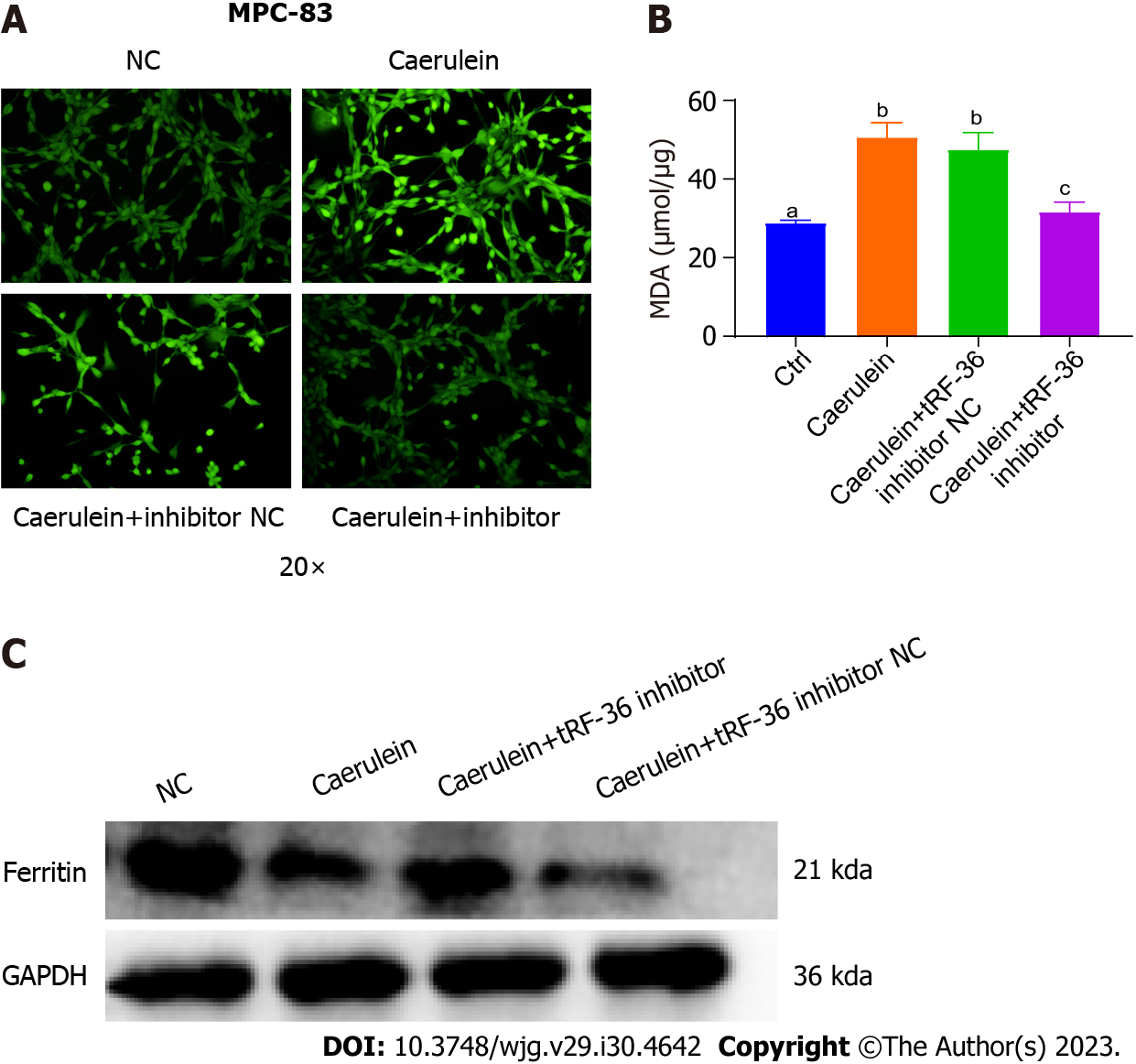
Figure 4 Effect of tRNA-derived fragments 36 on ferroptosis regulation in acute pancreatitis progression.
A: The reactive oxygen species levels were detected under a laser confocal microscope; B: Exploration of the malondialdehyde levels in the acute pancreatitis cell model; C: Ferritin levels using western blot analysis. a, b, and c represent different expression levels, the same letter represents no significant difference between groups (P > 0.05), different letters represent significant difference between groups (P < 0.05). tRF-36: tRNA-derived fragments 36; NC: Normal control.
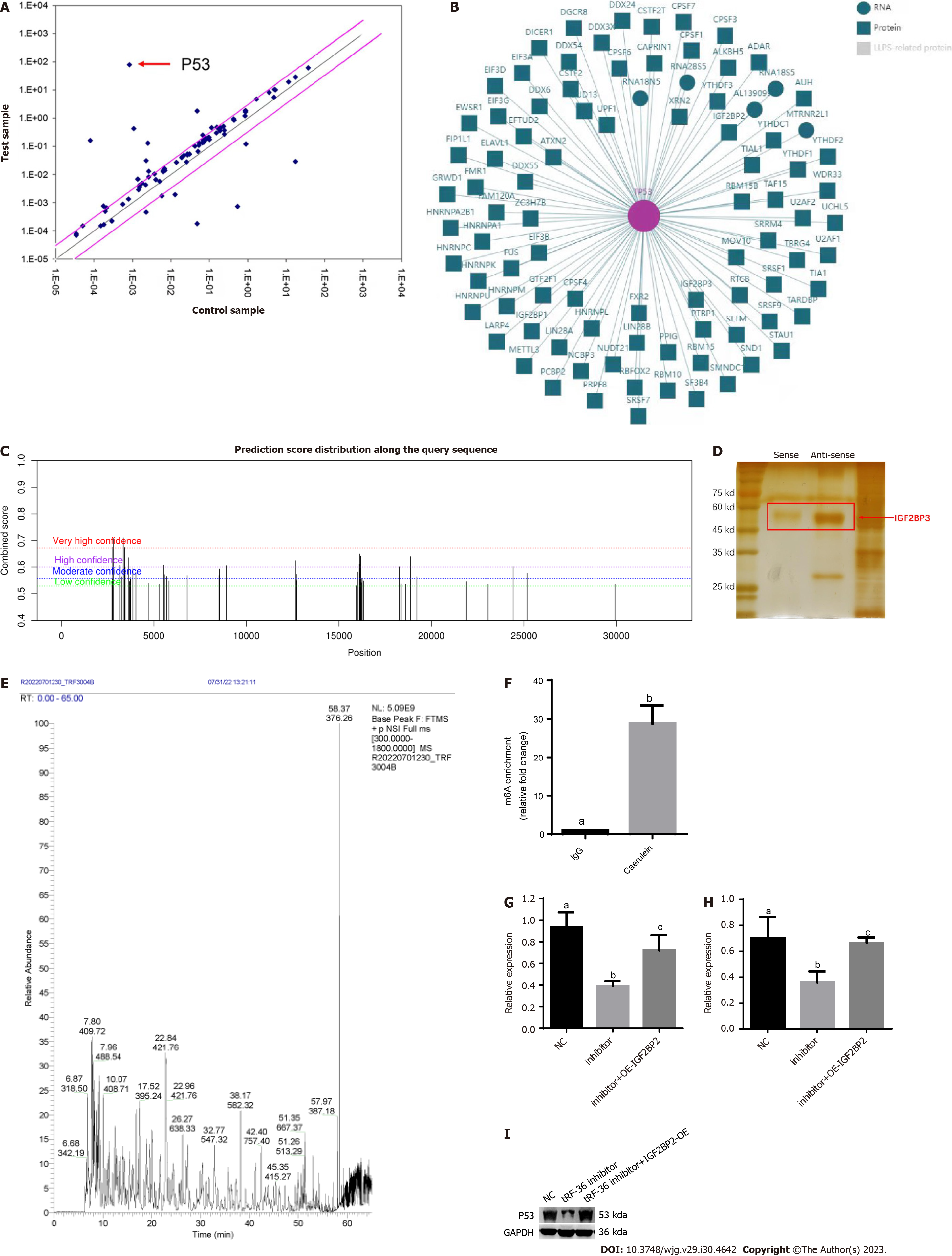
Figure 5 Exploration of the molecular mechanisms of tRNA-derived fragments 36 in acute pancreatitis progression.
A: p53 was the most significantly downregulated gene in ferroptosis gene microarray analysis after tRNA-derived fragments 36 (tRF-36) knockdown; B: The interaction network of p53 and m6A proteins; C: The m6A modification analysis for p53 mRNA by SRAMP; D: The RNA pull down assay for tRF-36 with insulin-like growth factor 2 mRNA binding protein 3; E: Mass spectrometry results for tRF-36; F: The level of m6A modification of p53 by methylated RNA immunoprecipitation-quantitative polymerase chain reaction.; G: The p53 expression level after the knockdown of tRF-36 by reverse transcription quantitative polymerase chain reaction; H: The p53 expression level after the knockdown of tRF-36 by western blot; I: The changes in P53 expression. a, b, and c represent different expression levels, the same letter represents no significant difference between groups (P > 0.05), different letters represent significant difference between groups (P < 0.05). tRF-36: tRNA-derived fragments 36; NC: Normal control; IgG: Immunoglobulin G; OE: Overexpression; IGF2BP3: Insulin-like growth factor 2 mRNA binding protein 3.
Figure 6 A schematic of a putative mechanism of tRNA-derived fragments 36 in the progression of acute pancreatitis.
In this model, tRNA-derived fragments 36 might recruit insulin-like growth factor 2 mRNA binding protein 3 (IGF2BP3) to the p53 mRNA m6A modification site by binding to IGF2BP3 to enhance p53 mRNA stability and promote the ferroptosis of pancreatic follicle cells. tRF-36: tRNA-derived fragments 36; IGF2BP3: Insulin-like growth factor 2 mRNA binding protein 3.
- Citation: Fan XR, Huang Y, Su Y, Chen SJ, Zhang YL, Huang WK, Wang H. Exploring the regulatory mechanism of tRNA-derived fragments 36 in acute pancreatitis based on small RNA sequencing and experiments. World J Gastroenterol 2023; 29(30): 4642-4656
- URL: https://www.wjgnet.com/1007-9327/full/v29/i30/4642.htm
- DOI: https://dx.doi.org/10.3748/wjg.v29.i30.4642