Copyright
©The Author(s) 2022.
World J Gastroenterol. Feb 28, 2022; 28(8): 825-839
Published online Feb 28, 2022. doi: 10.3748/wjg.v28.i8.825
Published online Feb 28, 2022. doi: 10.3748/wjg.v28.i8.825
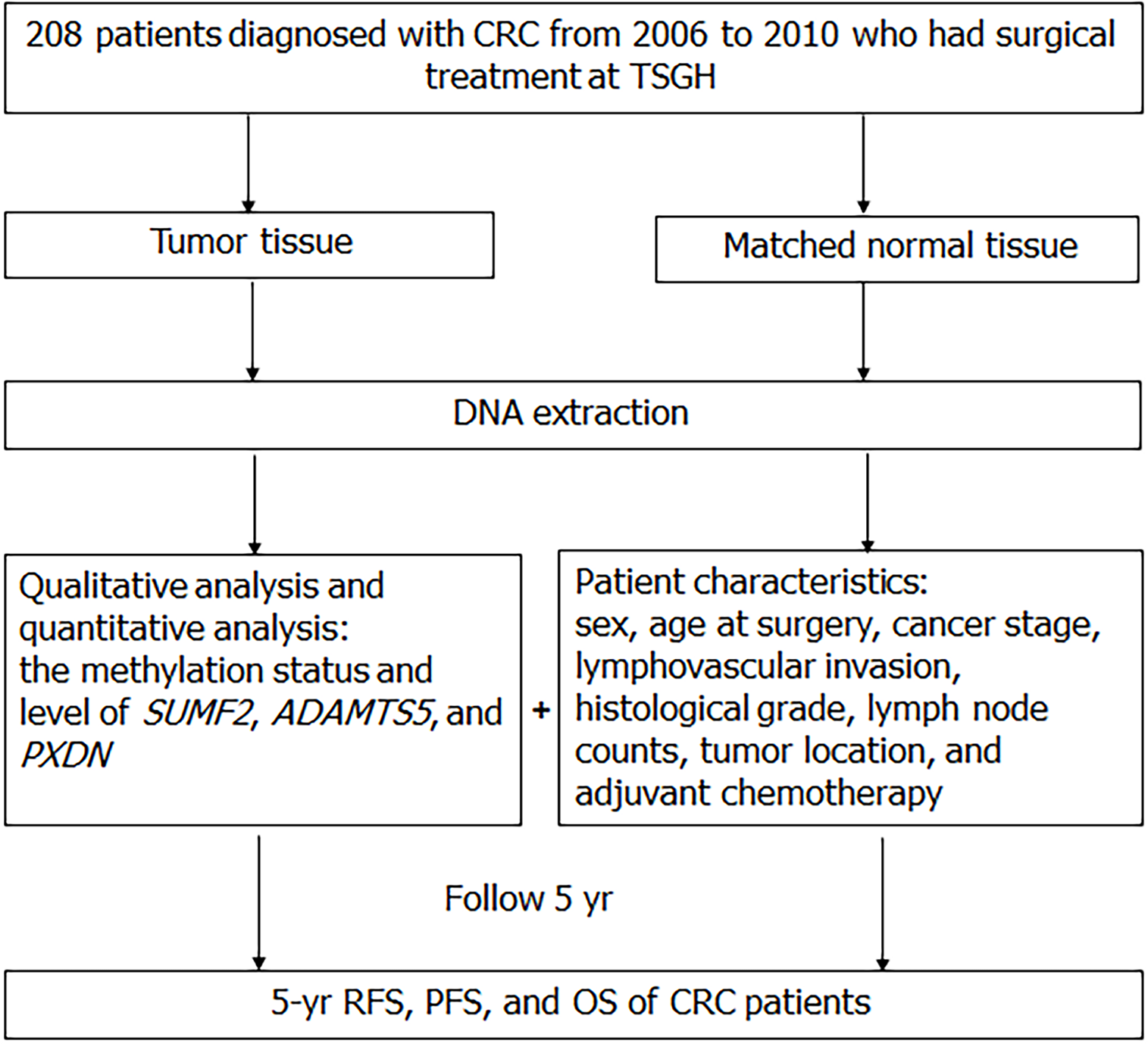
Figure 1 The study design flow-diagram.
CRC: Colorectal cancer; TSGH: Tri-Service General Hospital; SUMF2: Sulfatase modifying factor 2; ADAMTS5: ADAM metallopeptidase with thrombospondin type 1 motif 5; PXDN: Peroxidasin; RFS: Recurrence-free survival; PFS: Progression-free survival; OS: Overall survival.
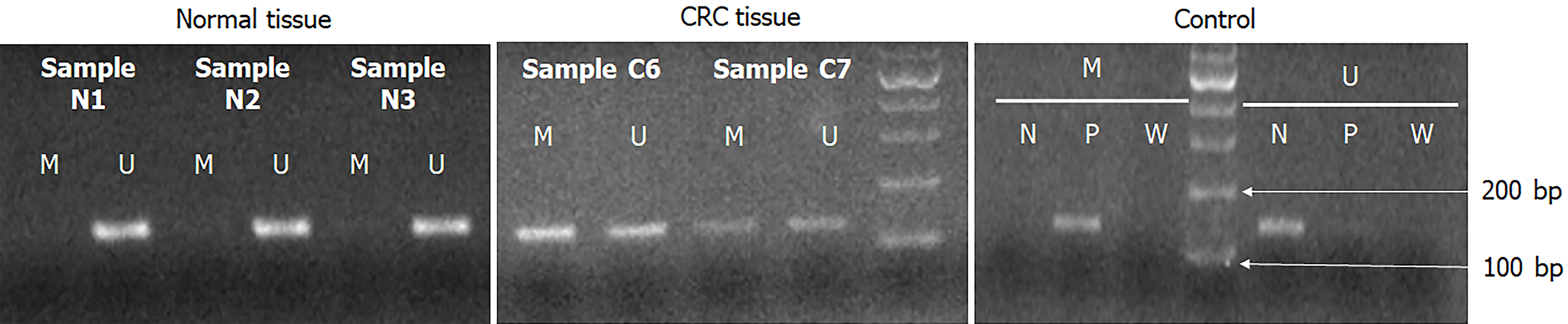
Figure 2 The methylation-specific polymerase chain reaction results of PXDN gene in colorectal cancer patients.
Negative control, positive control and sterile water represent unmethylation-specific reaction, methylation-specific reaction and no contaminant reaction for polymerase chain reaction, respectively. CRC: Colorectal cancer.
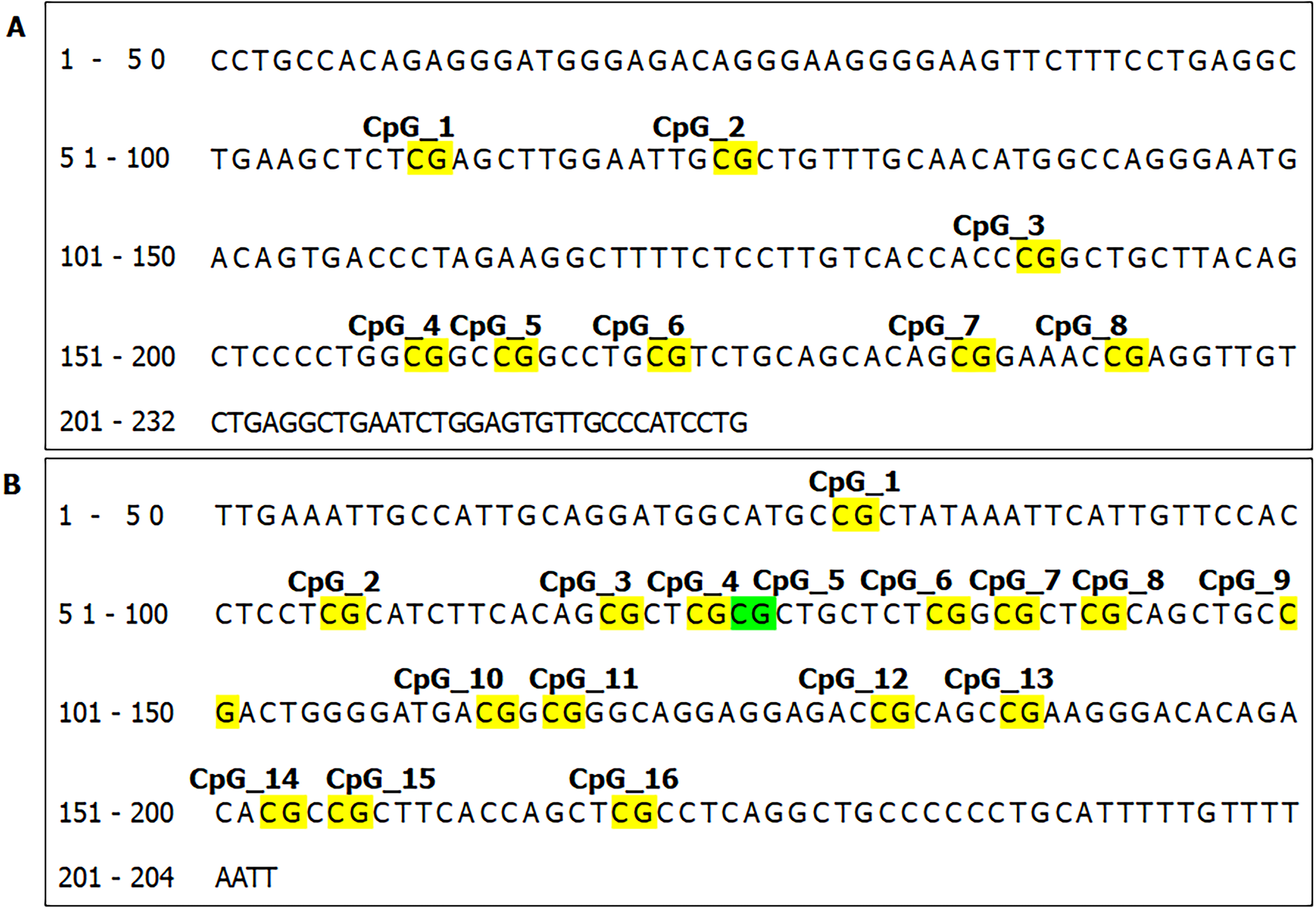
Figure 3 The location of informative CpG sites in the (A) SUMF2 and (B) ADAMTS5 promoter region.
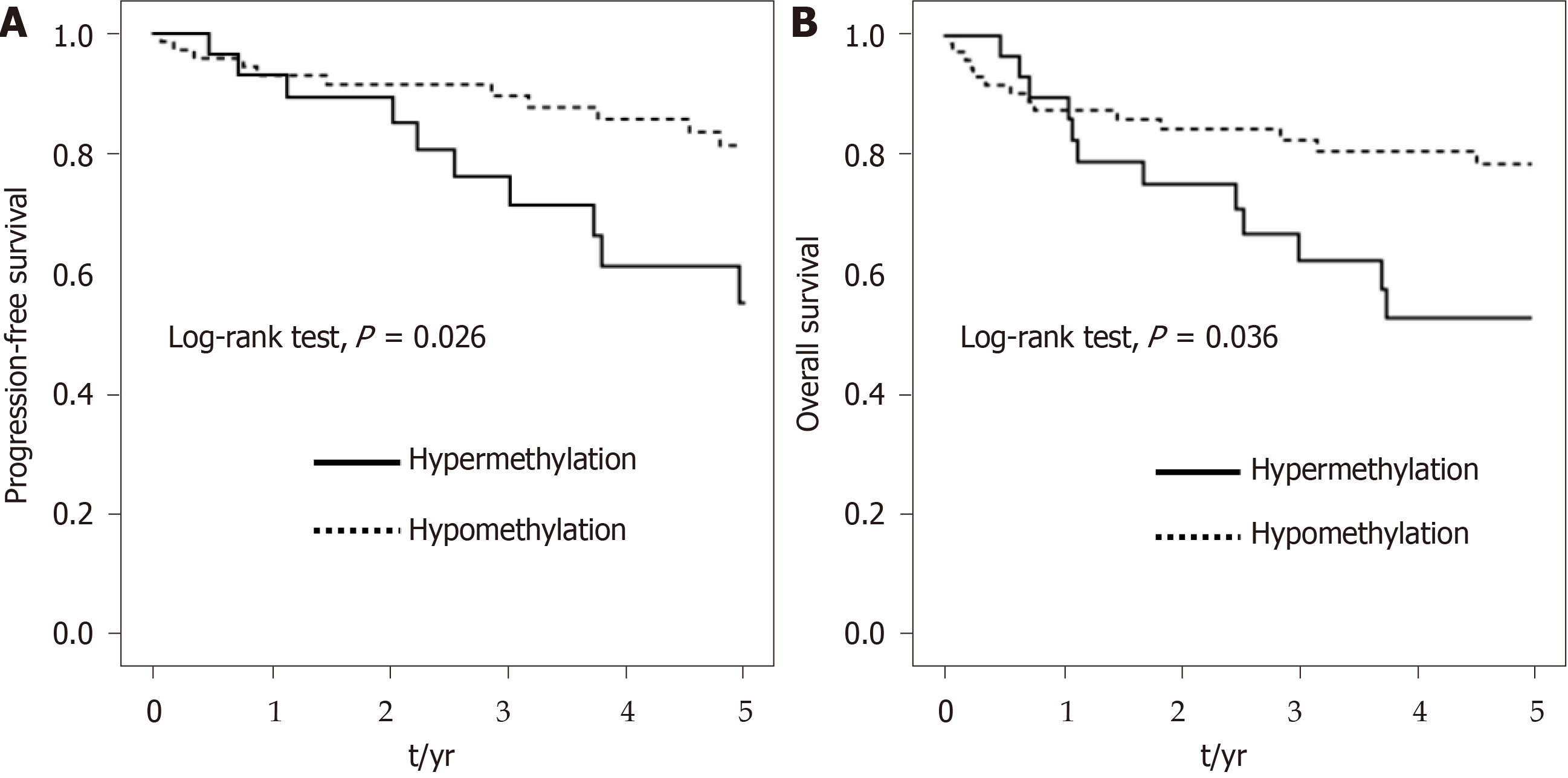
Figure 4 Kaplan–Meier survival curves depicting the effect of hypermethylation and hypomethylation of SUMF2 at CpG_3+CpG_7 from tumor tissue on 5-year (A) progression-free survival and (B) overall survival of colorectal cancer patients.
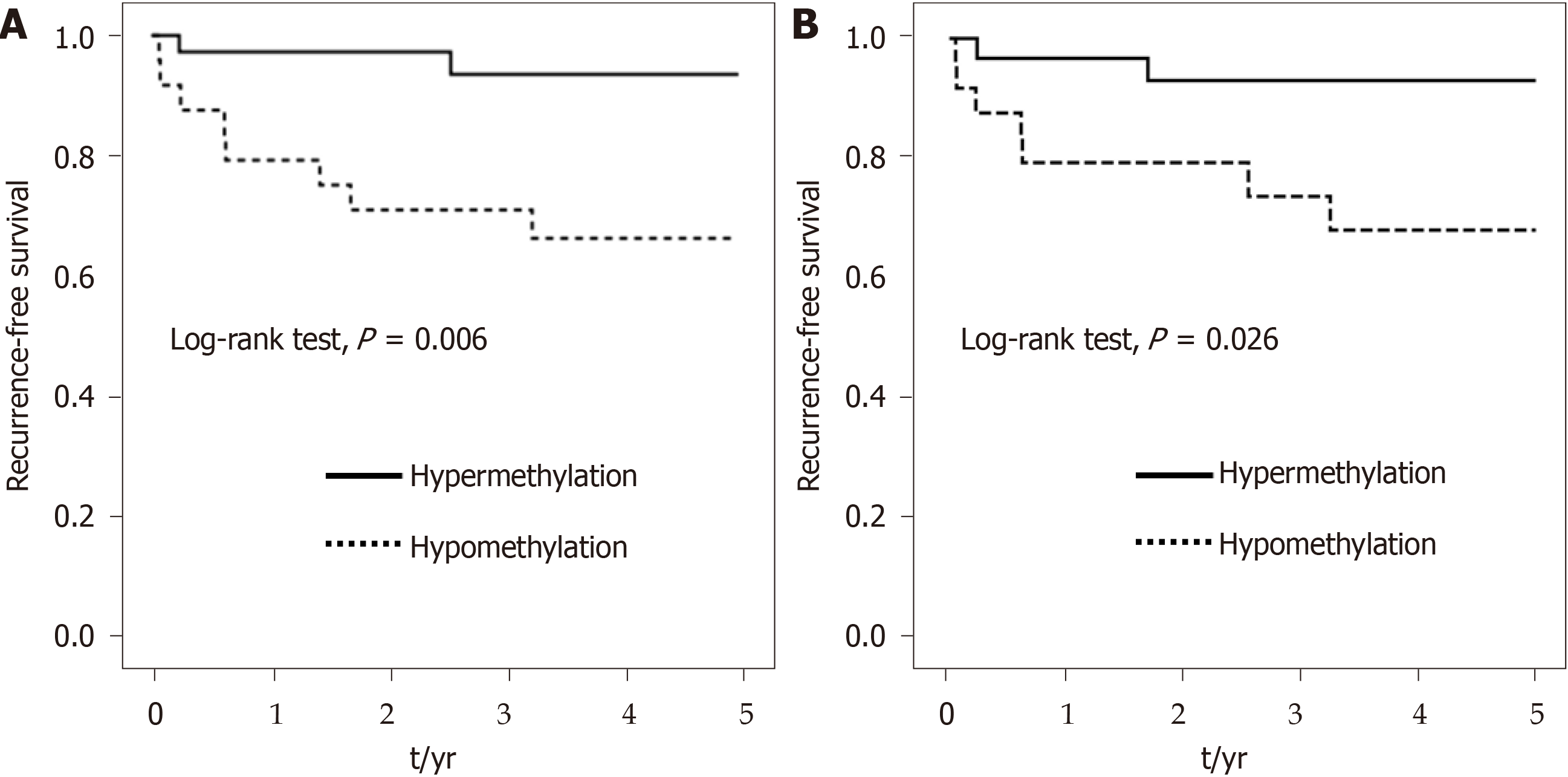
Figure 5 Kaplan–Meier survival curves depicting the effect of hypermethylation and hypomethylation of ADAMTS5 at (A) CpG_2 and (B) CpG_13 from normal tissue on 5-year recurrence-free survival of colorectal cancer patients.
- Citation: Su JQ, Lai PY, Hu PH, Hu JM, Chang PK, Chen CY, Wu JJ, Lin YJ, Sun CA, Yang T, Hsu CH, Lin HC, Chou YC. Differential DNA methylation analysis of SUMF2, ADAMTS5, and PXDN provides novel insights into colorectal cancer prognosis prediction in Taiwan. World J Gastroenterol 2022; 28(8): 825-839
- URL: https://www.wjgnet.com/1007-9327/full/v28/i8/825.htm
- DOI: https://dx.doi.org/10.3748/wjg.v28.i8.825