Copyright
©2014 Baishideng Publishing Group Inc.
World J Gastroenterol. Aug 7, 2014; 20(29): 10071-10081
Published online Aug 7, 2014. doi: 10.3748/wjg.v20.i29.10071
Published online Aug 7, 2014. doi: 10.3748/wjg.v20.i29.10071
Figure 1 KISS1 methylation analysis of colorectal cancer tumor samples by methylation-specific polymerase chain reaction.
Positive control: Placental tissue DNA treated with M.SssI methyltransferase; Negative control: DNA from normal lymphocytes. Tumors A and B were scored as positive/methylated due to the presence of a methylation band, whereas normal samples A and B were scored as negative/unmethylated due to the absence of a methylation band. The methylated polymerase chain reaction (PCR) product is 172 bp, and the unmethylated PCR product is 173 bp.
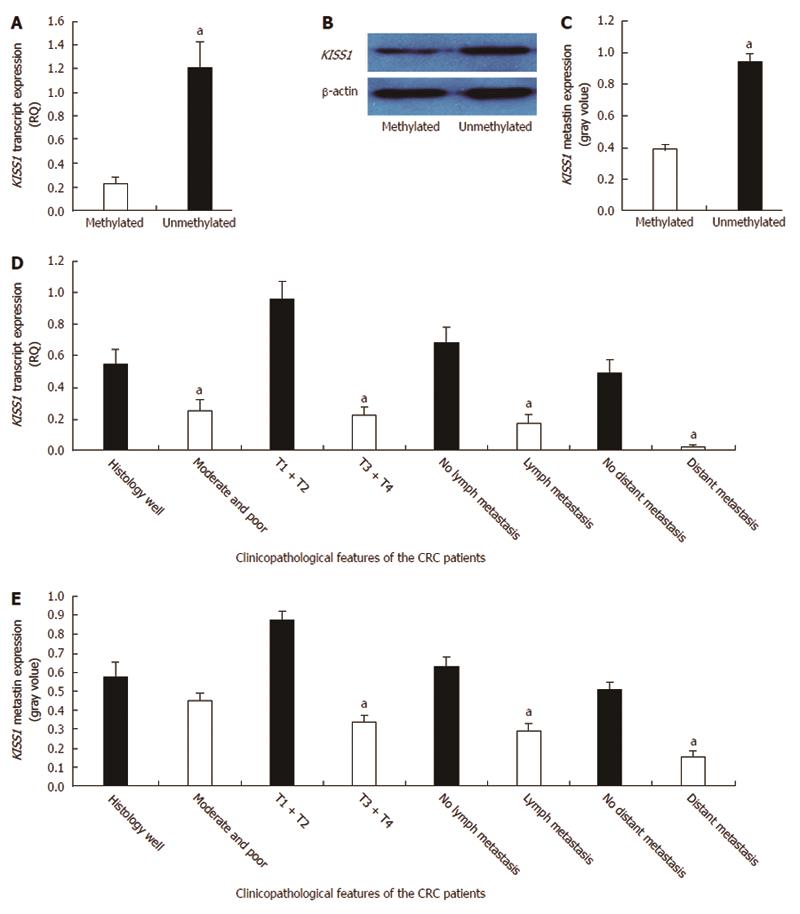
Figure 2 Correlations between KISS1 expression and methylation and clinicopathological factors in colorectal cancer patients.
A: KISS1 transcript expression in 126 colorectal cancer (CRC) patients was determined by real-time quantitative polymerase chain reaction (PCR). The data are presented as the mean ± SD. KISS1 transcript expression was significantly lower in the methylated KISS1 group than in the unmethylated KISS1 group (aP < 0.05 vs the methylated KISS1 group); B: KISS1/metastin protein levels in CRC tissues of the indicated genotypes were determined by Western blot. The same membranes were subsequently probed with an anti-β-actin antibody as a loading control; C: The KISS1/metastin protein expression levels are presented as the mean ± SEM. aP < 0.05 vs the methylated KISS1 group; D: The analysis of the relationship between KISS1 transcript expression and patient clinicopathological factors revealed a negative correlation with tumor differentiation, depth of invasion, lymph node metastasis and distant metastasis (aP < 0.05); E: Metastin expression was also negatively correlated with the depth of invasion, lymph node metastasis and distant metastasis (aP < 0.05).
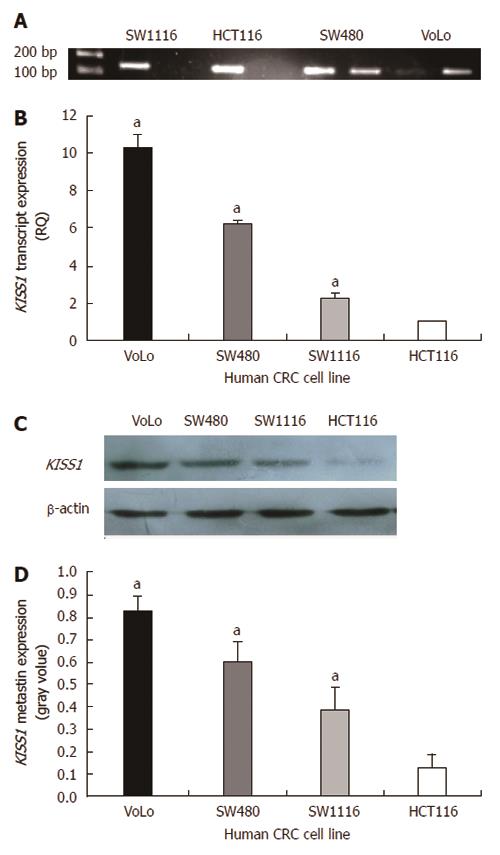
Figure 3 KISS1 methylation and KISS1 expression in various human colorectal cancer cell lines.
A: MS-PCR for KISS1 in human colorectal cancer (CRC) cell lines. A PCR band in lane M indicates a methylated KISS1 gene, whereas a band in lane U indicates an unmethylated KISS1 gene; B: KISS1 gene expression in CRC cell lines was examined by real-time PCR, and the expression of KISS1 was normalized to that of β-actin. The ratio of KISS1 expression in other CRC cells to KISS1 expression in HCT116 cells is shown on the Y-axis; C: Metastin expression in CRC cells of the indicated genotypes was determined by Western blot. The same membranes were subsequently probed with an anti-β-actin antibody as a loading control; D: The metastin expression levels are presented as the mean ± SEM. aP < 0.05 vs HCT116 cells.
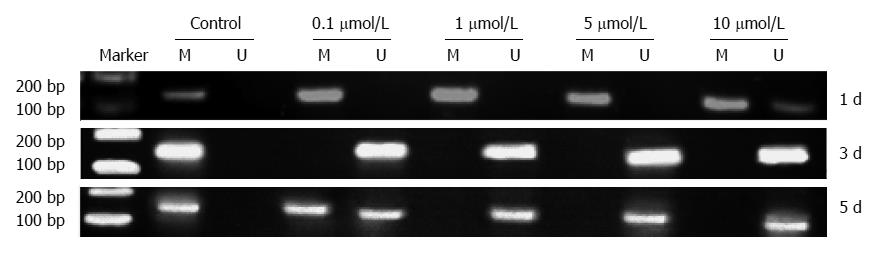
Figure 4 Changes in KISS1 methylation after treatment of HCT116 cells with different doses of 5-Aza-CdR.
MS-polymerase chain reaction (PCR) for KISS1 in HCT116 cells after 5-Aza-CdR treatment. HCT116 cells were treated for 1 d, 3 d or 5 d with 0 μmol/L (control), 0.1, 1, 5 or 10 μmol/L 5-Aza-CdR. A PCR band in lane M indicates a methylated KISS1 gene; a band in lane U indicates an unmethylated KISS1 gene. Marker: 100-bp DNA ladder I; M: Methylated primer product; U: Unmethylated primer product.
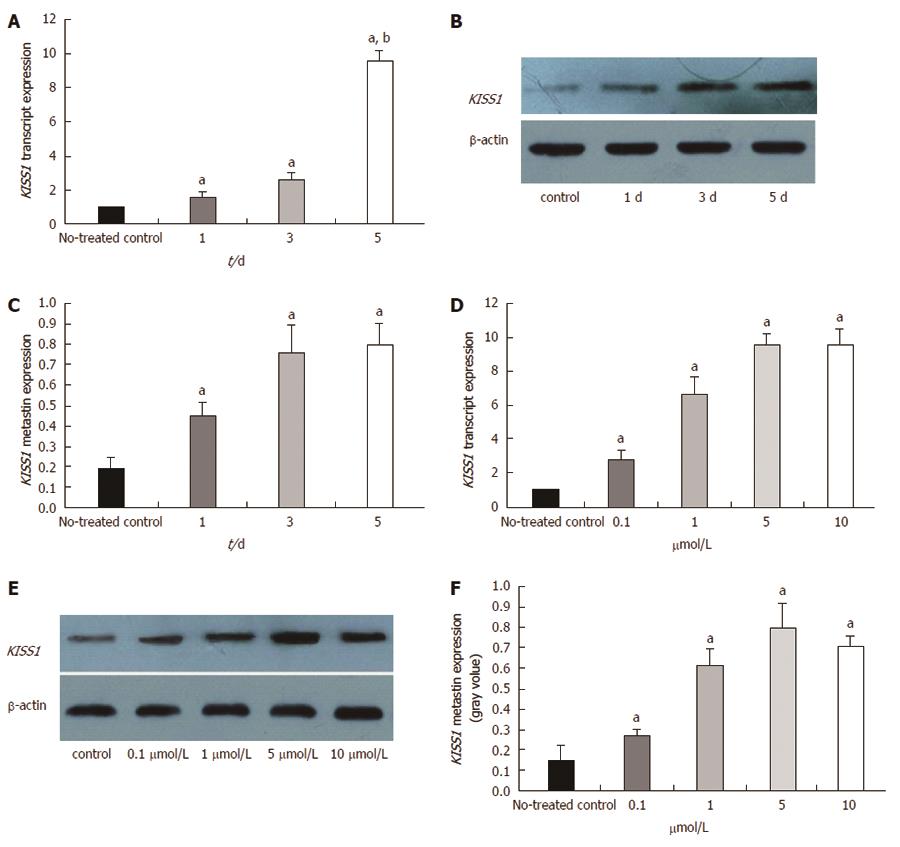
Figure 5 Changes in KISS1 expression after treatment of HCT116 cells with 5-Aza-CdR in vitro.
A: KISS1 transcript expression in HCT116 cells was examined by real-time polymerase chain reaction after 5-Aza-CdR treatment for the indicated times. KISS1 expression was normalized to that of β-actin. The ratio of KISS1 expression in treated cells to that in untreated control cells is plotted on the Y-axis. KISS1 transcript expression gradually increased over time; B: Metastin expression levels after treatment with 5 μmol/L 5-Aza-CdR for different times were determined by Western blot. The same membranes were subsequently probed with an anti-β-actin antibody as a loading control; C: Metastin expression levels gradually increased over time; D: KISS1 transcript expression gradually increased with increasing concentrations of 5-Aza-CdR after 5 d; E: Metastin protein levels were determined by Western blotting after a 5-d treatment with different concentrations of 5-Aza-CdR; F: Metastin expression gradually increased with increasing concentrations of 5-Aza-CdR. The data are presented as the mean ± SE. aP < 0.05 vs untreated control.
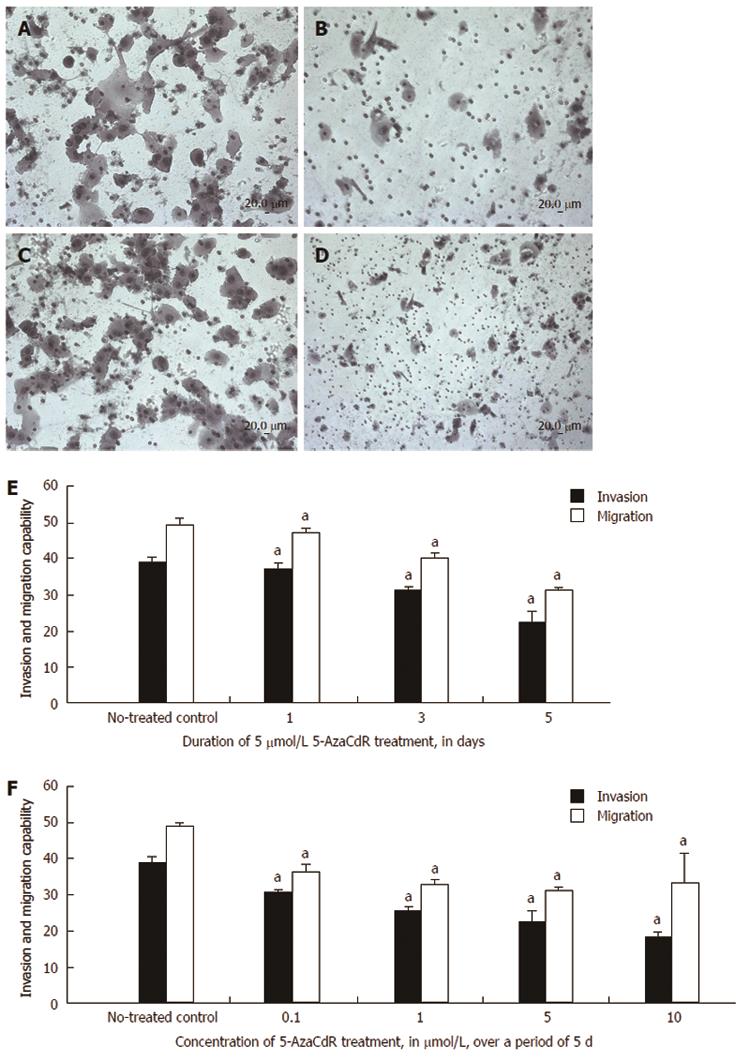
Figure 6 Changes in HCT116 cell invasion and migration after 5-Aza-CdR treatment.
A: The invasion of untreated HCT116 cells was determined using a Matrigel invasion assay; B: HCT116 cell invasion in one well after treatment with 5-Aza-CdR; C: Cell migration was determined in untreated cells using a migration assay; D: Cell migration after treatment with 5-Aza-CdR; E: HCT116 cell invasion and migration gradually decreased in a time-dependent manner after treatment; F: HCT116 cell invasion and migration gradually declined with increasing concentrations of 5-Aza-CdR. Each bar represents the mean of 3 independent wells. Scale bar, 20 μm. aP < 0.05 vs non-treated control.
-
Citation: Chen SQ, Chen ZH, Lin SY, Dai QB, Fu LX, Chen RQ.
KISS1 methylation and expression as predictors of disease progression in colorectal cancer patients. World J Gastroenterol 2014; 20(29): 10071-10081 - URL: https://www.wjgnet.com/1007-9327/full/v20/i29/10071.htm
- DOI: https://dx.doi.org/10.3748/wjg.v20.i29.10071